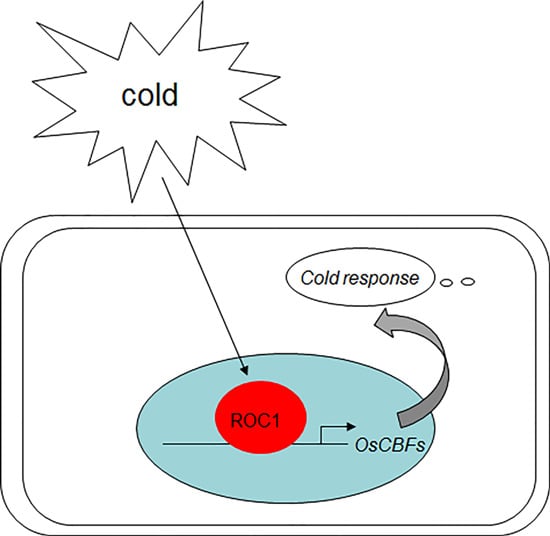
The Indeterminate Domain Protein ROC1 Regulates Chilling Tolerance via Activation of DREB1B/CBF1 in Rice
Abstract
:1. Introduction
2. Results
2.1. Isolation of the Regulators of CBF1

2.2. ROC1 Directly Binds to the Promoter of CBF1

2.3. ROC1 Is Localized in the Nucleus and Shows Trans-Activation Activity in Yeast

2.4. ROC1 Mutants Are Sensitive to Cold Stress

2.5. Cold-Induction of CBFs Is Inhibited in roc1 Mutants


3. Discussion
4. Materials and Methods
4.1. Mutant Isolation and Plant Growth
4.2. Plants Expressing the Plasmid Construction
4.3. Yeast One-Hybrid Analysis
4.4. Subcellular Localization
4.5. Trans-Activation Assays
4.6. RT-PCR Analysis
4.7. Northern Blot Analysis
4.8. Immunoblot Analysis
4.9. EMSA
4.10. ChIP Assay
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| IDD | Indeterminate domain |
| CBF | C-repeat binding factor |
| ROC1 | Regulator of CBF1 |
| ABA | Abscisic acid |
| NAA | 1-Naphthaleneacetic acid |
| BR | Brassinosteroid |
| GA | Gibberellic acid |
| ACC | 1-Amino-cyclopropane-1-carboxylic acid |
References
- Thomashow, M.F. PLANT COLD ACCLIMATION: Freezing Tolerance Genes and Regulatory Mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Khush, G.S. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 1997, 35, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Medina, J.; Bargues, M.; Terol, J.; Perez-Alonso, M.; Salinas, J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 1999, 119, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Zarka, D.G.; Stockinger, E.J.; Salazar, M.P.; Houghton, J.M.; Thomashow, M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998, 16, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef] [PubMed]
- Shinwari, Z.K.; Nakashima, K.; Miura, S.; Kasuga, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem. Biophys. Res. Commun. 1998, 250, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Kasuga, M.; Liu, Q.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999, 17, 287–291. [Google Scholar] [PubMed]
- Novillo, F.; Alonso, J.M.; Ecker, J.R.; Salinas, J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3985–3990. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.H.; Fujii, H.; Zheng, X.; Zhu, J.K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.K.; Sunkar, R. Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 2010, 639, 39–55. [Google Scholar] [PubMed]
- Dong, C.H.; Agarwal, M.; Zhang, Y.; Xie, Q.; Zhu, J.K. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 2006, 103, 8281–8286. [Google Scholar] [CrossRef] [PubMed]
- Ishitani, M.; Xiong, L.; Lee, H.; Stevenson, B.; Zhu, J.K. HOS1, a genetic locus involved in cold-responsive gene expression in arabidopsis. Plant Cell 1998, 10, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Gene Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Colasanti, J.; Yuan, Z.; Sundaresan, V. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 1998, 93, 593–603. [Google Scholar] [CrossRef]
- Park, S.J.; Kim, S.L.; Lee, S.; Je, B.I.; Piao, H.L.; Park, S.H.; Kim, C.M.; Ryu, C.H.; Park, S.H.; Xuan, Y.H.; et al. Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J. 2008, 56, 1018–1029. [Google Scholar] [CrossRef] [PubMed]
- Welch, D.; Hassan, H.; Blilou, I.; Immink, R.; Heidstra, R.; Scheres, B. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Gene Dev. 2007, 21, 2196–2204. [Google Scholar] [CrossRef] [PubMed]
- Feurtado, J.A.; Huang, D.; Wicki-Stordeur, L.; Hemstock, L.E.; Potentier, M.S.; Tsang, E.W.; Cutler, A.J. The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell 2011, 23, 1772–1794. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Ryu, J.; Kang, S.K.; Park, C.M. Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. Plant J. 2011, 65, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.T.; Sakaguchi, K.; Kiyose, S.; Taira, K.; Kato, T.; Nakamura, M.; Tasaka, M. A C2H2-type zinc finger protein, SGR5, is involved in early events of gravitropism in Arabidopsis inflorescence stems. Plant J. 2006, 47, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Tang, D.; Li, M.; Wang, K.; Cheng, Z. Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol. 2013, 161, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Xuan, Y.H.; Priatama, R.A.; Huang, J.; Je, B.I.; Liu, J.M.; Park, S.J.; Piao, H.L.; Son, D.Y.; Lee, J.J.; Park, S.H.; et al. Indeterminate domain 10 regulates ammonium-mediated gene expression in rice roots. New Phytol. 2013, 197, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Kozaki, A.; Hake, S.; Colasanti, J. The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res. 2004, 32, 1710–1720. [Google Scholar] [CrossRef] [PubMed]
- Baldoni, E.; Genga, A.; Medici, A.; Coraggio, I.; Locatelli, F. The OsMyb4 gene family: Stress response andtranscriptional auto-regulation mechanisms. Biol. Plant. 2013, 57, 691–700. [Google Scholar] [CrossRef]
- Agarwal, P.K.; Agarwal, P.; Reddy, M.K.; Sopory, S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006, 25, 1263–1274. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Liu, H.; Xiong, L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 2013, 4, 397. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, K.; Uemura, M.; Tsurumi, S.; Rahman, A. Auxin response in Arabidopsis under cold stress: Underlying molecular mechanisms. Plant Cell 2009, 21, 3823–3838. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Jin, J.B.; Lee, J.; Yoo, C.Y.; Stirm, V.; Miura, T.; Ashworth, E.N.; Bressan, R.A.; Yun, D.J.; Hasegawa, P.M. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 2007, 19, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.F.; Cai, W.H.; Jung, J.H.; Xuan, Y.H. NH4+-mediated protein phosphorylation in rice root. Acta Biol. Cracov. Bot. 2015, 57, 1–12. [Google Scholar]
- An, S.; Park, S.; Jeong, D.H.; Lee, D.Y.; Kang, H.G.; Yu, J.H.; Hur, J.; Kim, S.R.; Kim, Y.H.; Lee, M.; et al. Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol. 2003, 133, 2040–2047. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Jing, W.; Xiao, L.; Jin, Y.; Shen, L.; Zhang, W. The Rice High-Affinity Potassium Transporter1;1 Is Involved in Salt Tolerance and Regulated by an MYB-Type Transcription Factor. Plant Physiol. 2015, 168, 1076–1090. [Google Scholar] [CrossRef] [PubMed]
- James, P.; Halladay, J.; Craig, E.A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1996, 144, 1425–1436. [Google Scholar] [PubMed]
- Je, B.I.; Piao, H.L.; Park, S.J.; Park, S.H.; Kim, C.M.; Xuan, Y.H.; Park, S.H.; Huang, J.; Choi, Y.D.; An, G.; et al. RAV-Like1 maintains brassinosteroid homeostasis via the coordinated activation of BRI1 and biosynthetic genes in rice. Plant Cell 2010, 22, 1777–1791. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dou, M.; Cheng, S.; Zhao, B.; Xuan, Y.; Shao, M. The Indeterminate Domain Protein ROC1 Regulates Chilling Tolerance via Activation of DREB1B/CBF1 in Rice. Int. J. Mol. Sci. 2016, 17, 233. https://doi.org/10.3390/ijms17030233
Dou M, Cheng S, Zhao B, Xuan Y, Shao M. The Indeterminate Domain Protein ROC1 Regulates Chilling Tolerance via Activation of DREB1B/CBF1 in Rice. International Journal of Molecular Sciences. 2016; 17(3):233. https://doi.org/10.3390/ijms17030233
Chicago/Turabian StyleDou, Mingzhu, Shuai Cheng, Baotian Zhao, Yuanhu Xuan, and Minglong Shao. 2016. "The Indeterminate Domain Protein ROC1 Regulates Chilling Tolerance via Activation of DREB1B/CBF1 in Rice" International Journal of Molecular Sciences 17, no. 3: 233. https://doi.org/10.3390/ijms17030233
APA StyleDou, M., Cheng, S., Zhao, B., Xuan, Y., & Shao, M. (2016). The Indeterminate Domain Protein ROC1 Regulates Chilling Tolerance via Activation of DREB1B/CBF1 in Rice. International Journal of Molecular Sciences, 17(3), 233. https://doi.org/10.3390/ijms17030233