Structure of the PLP-Form of the Human Kynurenine Aminotransferase II in a Novel Spacegroup at 1.83 Å Resolution
Abstract
:1. Introduction
2. Results and Discussion
2.1. Purification and Activity Assay of hKAT-II
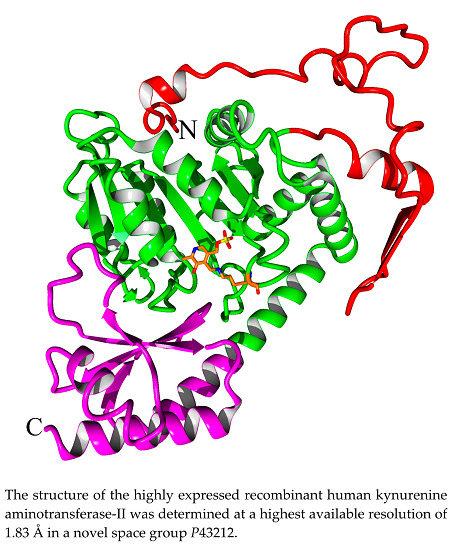
2.2. Overall Structure
3. Materials and Methods
3.1. Enzyme Expression and Purification
3.2. Activity Assay
3.3. Crystallization, Data Collection, Structure Solution and Refinement
3.4. Homology Modeling
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| KAT-II | Kynurenine aminotransferase II (KAT-II) |
| KYNA | Kynurenic acid |
| NMDA | N-methyl-d-aspartate |
| AMPA | α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid |
| α7-nAChRs | α7-nicotinic acetylcholine receptors |
| CNS | Central nervous system |
| L-KYN | l-kynurenine |
| PLP | Pyridoxal-5′-phosphate |
| 6xHis | Hexa-histidine |
| Ni-NTA | Nickel-nitrilotriacetic acid |
| IPTG | Isopropyl β-d-1-thiogalactopyranoside |
| DPI | Diffraction precision index |
References
- Battaglia, G.; Rassoulpour, A.; Wu, H.Q.; Hodgkins, P.S.; Kiss, C.; Nicoletti, F.; Schwarcz, R. Some metabotropic glutamate receptor ligands reduce kynurenate synthesis in rats by intracellular inhibition of kynurenine aminotransferase II. J. Neurochem. 2000, 75, 2051–2060. [Google Scholar] [CrossRef] [PubMed]
- Palmada, M.; Centelles, J.J. Excitatory amino acid neurotransmission. Pathways for metabolism, storage and reuptake of glutamate in brain. Front. Biosci. 1998, 3, d701–d718. [Google Scholar] [PubMed]
- Martin, S.J.; Grimwood, P.D.; Morris, R.G. Synaptic plasticity and memory: An evaluation of the hypothesis. Annu. Rev. Neurosci. 2000, 23, 649–711. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Robinson, H.; Li, J. Crystal structure of human kynurenine aminotransferase II. J. Biol. Chem. 2008, 283, 3567–3573. [Google Scholar] [CrossRef] [PubMed]
- Chon, H.; Matsumura, H.; Koga, Y.; Takano, K.; Kanaya, S. Crystal structure of a human kynurenine aminotransferase II homologue from Pyrococcus horikoshii OT3 at 2.20 Å resolution. Proteins 2005, 61, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Andine, P.; Lehmann, A.; Ellren, K.; Wennberg, E.; Kjellmer, I.; Nielsen, T.; Hagberg, H. The excitatory amino acid antagonist kynurenic acid administered after hypoxic-ischemia in neonatal rats offers neuroprotection. Neurosci. Lett. 1988, 90, 208–212. [Google Scholar] [CrossRef]
- Foster, A.C.; Vezzani, A.; French, E.D.; Schwarcz, R. Kynurenic acid blocks neurotoxicity and seizures induced in rats by the related brain metabolite quinolinic acid. Neurosci. Lett. 1984, 48, 273–278. [Google Scholar] [CrossRef]
- Vecsei, L.; Szalardy, L.; Fulop, F.; Toldi, J. Kynurenines in the CNS: Recent advances and new questions. Nat. Rev. Drug Discov. 2013, 12, 64–82. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Simonavicius, N.; Wu, X.S.; Swaminath, G.; Reagan, J.; Tian, H.; Ling, L. Kynurenic acid as a ligand for orphan G protein-coupled receptor GPR35. J. Biol. Chem. 2006, 281, 22021–22028. [Google Scholar] [CrossRef] [PubMed]
- Mellor, A.L.; Munn, D.H. IDO expression by dendritic cells: Tolerance and tryptophan catabolism. Nat. Rev. Immunol. 2004, 4, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Schwarcz, R.; Bruno, J.P.; Muchowski, P.J.; Wu, H.Q. Kynurenines in the mammalian brain: When physiology meets pathology. Nat. Rev. Neurosci. 2012, 13, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Oxenkrug, G.F. Tryptophan-Kynurenine Metabolism as a common mediator of genetic and environmental impacts in major depressive disorder: The serotonin hypothesis revisited 40 years later. Isr. J. Psychiatry Relat. Sci. 2010, 47, 56–63. [Google Scholar] [PubMed]
- Stone, T.W.; Darlington, L.G. Endogenous kynurenines as targets for drug discovery and development. Nat. Rev. Drug Discov. 2002, 1, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Lugo-Huitron, R.; Blanco-Ayala, T.; Ugalde-Muniz, P.; Carrillo-Mora, P.; Pedraza-Chaverri, J.; Silva-Adaya, D.; Maldonado, P.D.; Torres, I.; Pinzon, E.; Ortiz-Islas, E.; et al. On the antioxidant properties of kynurenic acid: Free radical scavenging activity and inhibition of oxidative stress. Neurotoxicol. Teratol. 2011, 33, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Hardeland, R.; Zsizsik, B.K.; Poeggeler, B.; Fuhrberg, B.; Holst, S.; Coto-Montes, A. Indole-3-pyruvic and -propionic acids, kynurenic acid, and related metabolites as luminophores and free-radical scavengers. Adv. Exp. Med. Biol. 1999, 467, 389–395. [Google Scholar] [PubMed]
- Szalardy, L.; Zadori, D.; Toldi, J.; Fulop, F.; Klivenyi, P.; Vecsei, L. Manipulating kynurenic acid levels in the brain—On the edge between neuroprotection and cognitive dysfunction. Curr. Top. Med. Chem. 2012, 12, 1797–1806. [Google Scholar] [CrossRef] [PubMed]
- Erhardt, S.; Blennow, K.; Nordin, C.; Skogh, E.; Lindstrom, L.H.; Engberg, G. Kynurenic acid levels are elevated in the cerebrospinal fluid of patients with schizophrenia. Neurosci. Lett. 2001, 313, 96–98. [Google Scholar] [CrossRef]
- Nematollahi, A.; Aminimoghadamfarouj, N.; Church, W.B. Essential structural features of novel antischizophrenic drugs: A review. Med. Chem. 2014, 10, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Nematollahi, A.; Church, W.B.; Nadvi, N.A.; Gorrell, M.D.; Sun, G. Homology modeling of human kynurenine aminotransferase III and observations on inhibitor binding using molecular docking. Cent. Nerv. Syst. Agents Med. Chem. 2014, 14, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Jayawickrama, G.S.; Sadig, R.R.; Sun, G.; Nematollahi, A.; Nadvi, N.A.; Hanrahan, J.R.; Gorrell, M.D.; Church, W.B. Kynurenine aminotransferases and the prospects of inhibitors for the treatment of schizophrenia. Curr. Med. Chem. 2015, 22, 2902–2918. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Cai, T.; Tagle, D.A.; Li, J. Structure, expression, and function of kynurenine aminotransferases in human and rodent brains. Cell. Mol. Life Sci. 2010, 67, 353–68. [Google Scholar] [CrossRef] [PubMed]
- Guidetti, P.; Hoffman, G.E.; Melendez-Ferro, M.; Albuquerque, E.X.; Schwarcz, R. Astrocytic localization of kynurenine aminotransferase II in the rat brain visualized by immunocytochemistry. Glia 2007, 55, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; di Prospero, N.A.; Sapko, M.T.; Cai, T.; Chen, A.; Melendez-Ferro, M.; Du, F.; Whetsell, W.O., Jr.; Guidetti, P.; Schwarcz, R.; et al. Biochemical and phenotypic abnormalities in kynurenine aminotransferase II-deficient mice. Mol. Cell. Biol. 2004, 24, 6919–6930. [Google Scholar] [CrossRef] [PubMed]
- Okuno, E.; Nakamura, M.; Schwarcz, R. Two kynurenine aminotransferases in human brain. Brain Res. 1991, 542, 307–312. [Google Scholar] [CrossRef]
- Reyes Ocampo, J.; Lugo Huitron, R.; Gonzalez-Esquivel, D.; Ugalde-Muniz, P.; Jimenez-Anguiano, A.; Pineda, B.; Pedraza-Chaverri, J.; Rios, C.; Perez de la Cruz, V. Kynurenines with neuroactive and redox properties: Relevance to aging and brain diseases. Oxid. Med. Cell. Longev. 2014, 2014, 646909. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.J.; Shurubor, Y.I.; Dorai, T.; Pinto, J.T.; Isakova, E.P.; Deryabina, Y.I.; Denton, T.T.; Krasnikov, B.F. Omega-Amidase: An underappreciated, but important enzyme in l-glutamine and l-asparagine metabolism; relevance to sulfur and nitrogen metabolism, tumor biology and hyperammonemic diseases. Amino Acids 2016, 48, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Stone, T.W. Endogenous neurotoxins from tryptophan. Toxicon 2001, 39, 61–73. [Google Scholar] [CrossRef]
- Jensen, R.A.; Gu, W. Evolutionary recruitment of biochemically specialized subdivisions of family I within the protein superfamily of aminotransferases. J. Bacteriol. 1996, 178, 2161–2171. [Google Scholar] [PubMed]
- Okamoto, A.; Kato, R.; Masui, R.; Yamagishi, A.; Oshima, T.; Kuramitsu, S. An aspartate aminotransferase from an extremely thermophilic bacterium, Thermus thermophilus HB8. J. Biochem. 1996, 119, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Guidetti, P.; Okuno, E.; Schwarcz, R. Characterization of rat brain kynurenine aminotransferases I and II. J. Neurosci. Res. 1997, 50, 457–465. [Google Scholar] [CrossRef]
- Goh, D.L.M.; Patel, A.; Thomas, G.H.; Salomons, G.S.; Schor, D.S.M.; Jakobs, C.; Geraghty, M.T. Characterization of the human gene encoding α-aminoadipate aminotransferase (AADAT). Mol. Genet. Metab. 2002, 76, 172–180. [Google Scholar] [CrossRef]
- Sun, G.; Nematollahi, A.; Nadvi, N.A.; Kwan, A.H.; Jeffries, C.M.; Church, W. Expression, purification and crystallization of human Kynurenine Aminotransferase 2 exploiting a highly optimized codon set. Protein Expr. Purif. 2016, 121, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Denesyuk, A.I.; Denessiouk, K.A.; Korpela, T.; Johnson, M.S. Functional attributes of the phosphate group binding cup of pyridoxal phosphate-dependent enzymes. J. Mol. Biol. 2002, 316, 155–172. [Google Scholar] [CrossRef] [PubMed]
- Denesyuk, A.I.; Denessiouk, K.A.; Korpela, T.; Johnson, M.S. Phosphate group binding “cup” of PLP-dependent and non-PLP-dependent enzymes: Leitmotif and variations. Biochim. Biophys. Acta 2003, 1647, 234–238. [Google Scholar] [CrossRef]
- Mehta, P.K.; Hale, T.I.; Christen, P. Aminotransferases—Demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 1993, 214, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Schneider, G.; Kack, H.; Lindqvist, Y. The manifold of vitamin B6 dependent enzymes. Structure 2000, 8, R1–R6. [Google Scholar] [CrossRef]
- Jansonius, J.N. Structure, evolution and action of vitamin B6-dependent enzymes. Curr. Opin. Struct. Biol. 1998, 8, 759–769. [Google Scholar] [CrossRef]
- Ku, S.Y.; Yip, P.; Howell, P.L. Structure of Escherichia coli tryptophanase. Acta Crystallogr. D Biol. Crystallogr. 2006, 62, 814–823. [Google Scholar] [CrossRef] [PubMed]
- Hasse, D.; Andersson, E.; Carlsson, G.; Masloboy, A.; Hagemann, M.; Bauwe, H.; Andersson, I. Structure of the homodimeric glycine decarboxylase P-protein from Synechocystis sp. PCC 6803 suggests a mechanism for redox regulation. J. Biol. Chem. 2013, 288, 35333–35345. [Google Scholar] [CrossRef] [PubMed]
- Gurusaran, M.; Shankar, M.; Nagarajan, R.; Helliwell, J.R.; Sekar, K. Do we see what we should see? Describing non-covalent interactions in protein structures including precision. IUCrJ 2014, 1 Pt 1, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.S.D.; Gurusaran, M.; Satheesh, S.N.; Radha, P.; Pavithra, S.; Tharshan, K.P.S.T.; Helliwell, J.R.; Sekar, K. Online_DPI: A web server to calculate the diffraction precision index for a protein structure. J. Appl. Crystallogr. 2015, 48, 939–942. [Google Scholar] [CrossRef]
- McPhillips, T.M.; McPhillips, S.E.; Chiu, H.J.; Cohen, A.E.; Deacon, A.M.; Ellis, P.J.; Garman, E.; Gonzalez, A.; Sauter, N.K.; Phizackerley, R.P.; et al. Blu-ice and the distributed control system: Software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 2002, 9, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D 2010, 66, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.R.; Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. D 2013, 69, 1204–1214. [Google Scholar] [CrossRef] [PubMed]
- Mccoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Bunkoczi, G.; Read, R.J. Improvement of molecular-replacement models with sculptor. Acta Crystallogr. D 2011, 67, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Garavaglia, S.; Montalbano, V.; Walsh, M.A.; Rizzi, M. Crystal structure of human kynurenine aminotransferase II, a drug target for the treatment of schizophrenia. J. Biol. Chem. 2008, 283, 3559–3566. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Cai, T.; Tagle, D.A.; Robinson, H.; Li, J.Y. Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II. Biosci. Rep. 2008, 28, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Valentina, C.; Garavaglia, S.; Sathyasaikumar, K.V.; Schwarcz, R.; Kojima, S.; Okuwaki, K.; Ono, S.; Kajii, Y.; Rizzi, M. Crystal structure-based selective targeting of the pyridoxal 5′-phosphate dependent enzyme kynurenine aminotransferase ii for cognitive enhancement. J. Med. Chem. 2010, 53, 5684–5689. [Google Scholar] [CrossRef] [PubMed]
- Tuttle, J.B.; Anderson, M.; Bechle, B.M.; Campbell, B.M.; Chang, C.; Dounay, A.B.; Evrard, E.; Fonseca, K.R.; Gan, X.M.; Ghosh, S.; et al. Structure-based design of irreversible human KAT II inhibitors: Discovery of new potency-enhancing interactions. ACS Med. Chem. Lett. 2013, 4, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Dounay, A.B.; Anderson, M.; Bechle, B.M.; Evrard, E.; Gan, X.M.; Kim, J.Y.; McAllister, L.A.; Pandit, J.; Rong, S.B.; Salafia, M.A.; et al. PF-04859989 as a template for structure-based drug design: Identification of new pyrazole series of irreversible KAT II inhibitors with improved lipophilic efficiency. Bioorg. Med. Chem. Lett. 2013, 23, 1961–1966. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Joo, K.; Lee, J.; Lee, J.; Raman, S.; Thompson, J.; Tyka, M.; Baker, D.; Karplus, K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins 2009, 77, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Davis, I.W.; Richardson, D.C. KiNG (Kinemage, Next Generation): A versatile interactive molecular and scientific visualization program. Protein Sci. 2009, 18, 2403–2409. [Google Scholar] [CrossRef] [PubMed]
- Lovell, S.C.; Word, J.M.; Richardson, J.S.; Richardson, D.C. The penultimate rotamer library. Proteins 2000, 40, 389–408. [Google Scholar] [CrossRef]
- Davis, I.W.; Arendall, W.B.; Richardson, D.C.; Richardson, J.S. The backrub motion: How protein backbone shrugs when a sidechain dances. Structure 2006, 14, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Urzhumtseva, L.; Afonine, P.V.; Adams, P.D.; Urzhumtsev, A. Crystallographic model quality at a glance. Acta Crystallogr. D 2009, 65, 297–300. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific LLC: San Carlos, CA, USA, 2002. [Google Scholar]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- King, R.D.; Sternberg, M.J.E. Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci. 1996, 5, 2298–2310. [Google Scholar] [CrossRef] [PubMed]




| PDB Entry Code | DPI * (Å) |
|---|---|
| 5EUN (structure reported here) | 0.12 |
| 2R2N | 0.20 |
| 2QLR | 0.50 |
| 2VGZ | 0.31 |
| 3DC1 | 0.32 |
| Source Organism | Homo Sapiens |
|---|---|
| DNA source | Synthesized (GenScript, Piscataway, NJ, USA) |
| 5′ sequence | BamHI/NdeI site: GGATCCCATATG |
| 3′ sequence | Stop codon and BamHI site: TAATAAGGATCC |
| Cloning vector | pUC57 |
| Expression vector | pET15b |
| Expression host | E. coli Rosetta 2 |
| Crystallization Method | Hanging Drop Vapor Diffusion |
|---|---|
| Plate type | 24-well Tissue plate |
| Temperature (K) | 293.15 |
| Protein concentration (mg/mL) | 7 |
| Buffer of protein solution | 20 mM Tris-HCl, pH 8.0, 50 mM NaCl |
| Reservoir solution | 200 mM NaCl, 0.1 M, NaCitrate pH 5.6, 24% PEG4K |
| Volume and ratio of drop | 2 μL, 1:1 ratio |
| Volume of reservoir (mL) | 1 |
| Diffraction Source | Australian Synchrotron MX2 |
|---|---|
| Wavelength (Å) | 0.9537 |
| Temperature (K) | 100 |
| Detector | ADSC QUANTUM 315r CCD |
| Crystal-to-detector distance (mm) | 250 |
| Rotation range per image (°) | 1.0 |
| Total rotation range (°) | 180 |
| Exposure time per image (s) | 1 |
| Space group | P43212 |
| Unit cell (Å, °) | a = b = 102.46, c = 86.24 (α, β and γ = 90) |
| Resolution (Å) | 39.74–1.825 (1.891–1.825) * |
| Total reflections | 81,444 (6884) |
| Unique reflections | 40,739 (3147) |
| Multiplicity | 2.0 (2.0) |
| Completeness (%) | 98.5 (85.1) |
| 〈I/σ(I)〉 | 17.64 (2.87) |
| Wilson B-factor | 17.44 |
| Rmerge | 0.0231 (0.1859) |
| Rmeas | 0.03267 (0.2629) |
| CC1/2 | 0.999 (0.909) |
| Reflections used in refinement | 39644 (3147) |
| Reflections used for Rfree | 1942 (152) |
| Rwork | 0.1730 (0.2148) |
| Rfree | 0.1988 (0.2696) |
| CCwork | 0.958 (0.906) |
| CCfree | 0.932 (0.780) |
| Protein residues | 425 |
| R.m.s. (bonds) | 0.004 |
| R.m.s (angles) | 0.72 |
| Average B-factor | 27.95 |
| Ramachandran favored (%) | 95.5 |
| Ramachandran allowed (%) | 4.5 |
| Ramachandran outliers (%) | 0 |
| Rotamer outliers (%) | 0.81 |
| Clashscore | 2.69 |
| PDB ID | 5EUN |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nematollahi, A.; Sun, G.; Harrop, S.J.; Hanrahan, J.R.; Church, W.B. Structure of the PLP-Form of the Human Kynurenine Aminotransferase II in a Novel Spacegroup at 1.83 Å Resolution. Int. J. Mol. Sci. 2016, 17, 446. https://doi.org/10.3390/ijms17040446
Nematollahi A, Sun G, Harrop SJ, Hanrahan JR, Church WB. Structure of the PLP-Form of the Human Kynurenine Aminotransferase II in a Novel Spacegroup at 1.83 Å Resolution. International Journal of Molecular Sciences. 2016; 17(4):446. https://doi.org/10.3390/ijms17040446
Chicago/Turabian StyleNematollahi, Alireza, Guanchen Sun, Stephen J. Harrop, Jane R. Hanrahan, and W. Bret Church. 2016. "Structure of the PLP-Form of the Human Kynurenine Aminotransferase II in a Novel Spacegroup at 1.83 Å Resolution" International Journal of Molecular Sciences 17, no. 4: 446. https://doi.org/10.3390/ijms17040446
APA StyleNematollahi, A., Sun, G., Harrop, S. J., Hanrahan, J. R., & Church, W. B. (2016). Structure of the PLP-Form of the Human Kynurenine Aminotransferase II in a Novel Spacegroup at 1.83 Å Resolution. International Journal of Molecular Sciences, 17(4), 446. https://doi.org/10.3390/ijms17040446