Structural Characterization of Heme Environmental Mutants of CgHmuT that Shuttles Heme Molecules to Heme Transporters
Abstract
:1. Introduction
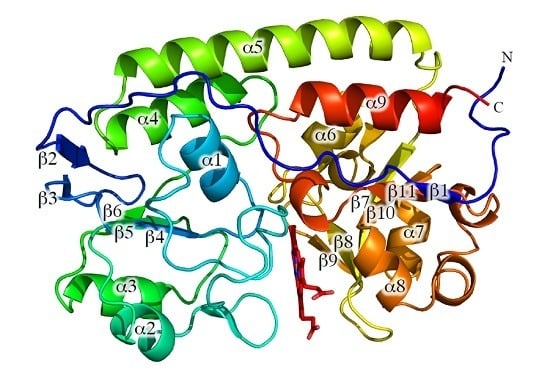
2. Results
3. Discussion
4. Materials and Methods
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ABC | ATP-Binding Cassette |
| ATPase | adenosine triphosphatase |
| CO | carbon monoxide |
| CT | charge transfer |
| 5cHS | five-coordinate high-spin |
| Hb | hemoglobin |
| Isd | iron-regulated surface determinant |
| NEAT | near iron transporter |
| r.m.s. | root-mean-square |
| 6cHS | six-coordinate high-spin |
| 6cLS | six-coordinate low-spin |
References
- Tong, Y.; Guo, M. Bacterial heme-transport proteins and their heme-coordination modes. Arch. Biochem. Biophys. 2009, 481, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Nobles, C.L.; Maresso, A.W. The theft of host heme by Gram-positive pathogenic bacteria. Metallomics 2011, 3, 788–796. [Google Scholar] [CrossRef] [PubMed]
- Hammer, N.D.; Skaar, E.P. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu. Rev. Microbiol. 2011, 65, 129–147. [Google Scholar] [CrossRef] [PubMed]
- Wandersman, C.; Delepelaire, P. Bacterial iron sources: From siderophores to hemophores. Annu. Rev. Microbiol. 2004, 58, 611–647. [Google Scholar] [CrossRef] [PubMed]
- Cescau, S.; Cwerman, H.; Létoffé, S.; Delepelaire, P.; Wandersman, C.; Biville, F. Heme acquisition by hemophores. Biometals 2007, 20, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Skaar, E.P.; Schneewind, O. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: Stealing iron from heme. Microbes Infect. 2004, 6, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Reniere, M.L.; Torres, V.J.; Skaar, E.P. Intracellular metalloporphyrin metabolism in Staphylococcus aureus. Biometals 2007, 20, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Grigg, J.C.; Ukpabi, G.; Gaudin, C.F.; Murphy, M.E. Structural biology of heme binding in the Staphylococcus aureus Isd system. J. Inorg. Biochem. 2010, 104, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Clarke, S.R.; Foster, S.J. Surface adhesins of Staphylococcus aureus. Adv. Microb. Physiol. 2006, 51, 187–224. [Google Scholar] [PubMed]
- Mazmanian, S.K.; Skaar, E.P.; Gasper, A.H.; Humayun, M.; Gornicki, P.; Jelenska, J.; Joachimiak, A.; Missiakas, D.M.; Schneewind, O. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 2003, 299, 906–909. [Google Scholar] [CrossRef] [PubMed]
- Andrade, M.A.; Ciccarelli., F.D.; Perez-Iratxeta, C.; Bork, P. NEAT: A domain duplicated in genes near the components of a putative Fe3+ siderophore transporter from Gram-positive pathogenic bacteria. Genome Biol. 2002, 3, 1–9. [Google Scholar] [CrossRef]
- Grigg, J.C.; Vermeiren, C.L.; Heinrichs, D.E.; Murphy, M.E. Heme coordination by Staphylococcus aureus IsdE. J. Biol. Chem. 2007, 282, 28815–22882. [Google Scholar] [CrossRef] [PubMed]
- Grigg, J.C.; Vermeiren, C.L.; Heinrichs, D.E.; Murphy, M.E. Haem recognition by a Staphylococcus aureus NEAT domain. Mol. Microbiol. 2007, 63, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Tanaka, W.N.; Zhu, H.; Xie, G.; Dooley, D.M.; Lei, B. Direct hemin transfer from IsdA to IsdC in the iron-regulated surface determinant (Isd) heme acquisition system of Staphylococcus aureus. J. Biol. Chem. 2008, 283, 6668–6676. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Xie, G.; Liu, M.; Olson, J.S.; Fabian, M.; Dooley, D.M.; Lei, B. Pathway for heme uptake from human methemoglobin by the iron-regulated surface determinants system of Staphylococcus aureus. J. Biol. Chem. 2008, 283, 18450–18460. [Google Scholar] [CrossRef] [PubMed]
- Muryoi, N.; Tiedemann, M.T.; Pluym, M.; Cheung, J.; Heinrichs, D.E.; Stillman, M.J. Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus. J. Biol. Chem. 2008, 283, 28125–28136. [Google Scholar] [CrossRef] [PubMed]
- Maresso, A.W.; Chapa, T.J.; Schneewind, O. Surface protein IsdC and Sortase B are required for heme-iron scavenging of Bacillus anthracis. J. Bacteriol. 2006, 188, 8145–8152. [Google Scholar] [CrossRef] [PubMed]
- Gat, O.; Zaide, G.; Inbar, I.; Grosfeld, H.; Chitlaru, T.; Levy, H.; Shafferman, A. Characterization of Bacillus anthracis iron-regulated surface determinant (Isd) proteins containing NEAT domains. Mol. Microbiol. 2008, 70, 983–999. [Google Scholar] [PubMed]
- Drazek, E.S.; Hammack, C.A.; Schmitt, M.P. Corynebacterium diphtheriae genes required for acquisition of iron from haemin and haemoglobin are homologous to ABC haemin transporters. Mol. Microbiol. 2000, 36, 68–84. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.E.; Schmitt, M.P. HtaA is an iron-regulated hemin binding protein involved in the utilization of heme iron in Corynebacterium diphtheriae. J. Bacteriol. 2009, 191, 2638–2648. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.E.; Schmitt, M.P. Novel hemin binding domains in the Corynebacterium diphtheriae HtaA protein interact with hemoglobin and are critical for heme iron utilization by HtaA. J. Bacteriol. 2011, 193, 5374–5385. [Google Scholar] [CrossRef] [PubMed]
- Aranda, R.; Worley, C.E.; Liu, M.; Bitto, E.; Cates, M.S.; Olson, J.S.; Lei, B.; Phillips, G.N. Bis-methionyl coordination in the crystal structure of the heme-binding domain of the streptococcal cell surface protein Shp. Biochemistry 2007, 374, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Bates, C.S.; Montañez, G.E.; Woods, C.R.; Vincent, R.M.; Eichenbaum, Z. Identification and characterization of a Streptococcus pyogenes operon involved in binding of hemoproteins and acquisition of iron. Infect. Immun. 2003, 71, 1042–1055. [Google Scholar] [CrossRef] [PubMed]
- Ouattara, M.; Cunha, E.B.; Li, X.; Huang, Y.S.; Dixon, D.; Eichenbaum, Z. Shr of group A streptococcus is a new type of composite NEAT protein involved in sequestering haem from methaemoglobin. Mol. Microbiol. 2010, 78, 739–756. [Google Scholar] [CrossRef] [PubMed]
- Lei, B.; Liu, M.; Voyich, J.M.; Prater, C.I.; Kala, S.V.; DeLeo, F.R.; Musser, J.M. Identification and characterization of HtsA, a second heme-binding protein made by Streptococcus pyogenes. Infect. Immun. 2003, 71, 5962–5969. [Google Scholar] [CrossRef] [PubMed]
- Muraki, N.; Aono, S. Structural basis for heme recognition by HmuT responsible for heme transport to the heme transporter in Corynebacterium glutamicum. Chem. Lett. 2016, 45, 24–26. [Google Scholar] [CrossRef]
- Ran, Y.; Liu, M.; Zhu, H.; Nygaard, T.K.; Brown, D.E.; Fabian, M.; Dooley, D.M.; Lei, B. Spectroscopic identification of heme axial ligands in HtsA that are involved in heme acquisition by Streptococcus pyogenes. Biochemistry 2010, 49, 2834–2842. [Google Scholar] [CrossRef] [PubMed]
- Dali Server. Available online: http://ekhidna.biocenter.helsinki.fi/dali_server (accessed on 26 May 2016).
- Mattle, D.; Zeltina, A.; Woo, J.S.; Goetz, B.A.; Locher, K.P. Two stacked heme molecules in the binding pocket of the periplasmic heme-binding protein HmuT from Yersinia pestis. J. Mol. Biol. 2010, 404, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Ho, W.W.; Li, H.; Eakanunkul, S.; Tong, Y.; Wilks, A.; Guo, M.; Poulos, T.L. Holo- and apo-bound structures of bacterial periplasmic heme-binding proteins. J. Biol. Chem. 2007, 282, 35796–35802. [Google Scholar] [CrossRef] [PubMed]
- Krewulak, K.D.; Vogel, H.J. Structural biology of bacterial iron uptake. Biochim. Biophys. Acta 2008, 1778, 1781–1804. [Google Scholar] [CrossRef] [PubMed]
- Tam, R.; Saier, M.H., Jr. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 1993, 57, 320–346. [Google Scholar] [PubMed]
- Bordignon, E.; Grote, M.; Schneider, E. The maltose ATP-binding cassette transporter in the 21st century—Towards a structural dynamic perspective on its mode of action. Mol. Microbiol. 2010, 77, 1354–1366. [Google Scholar] [CrossRef] [PubMed]
- Hvorup, R.N.; Goetz, B.A.; Niederer, M.; Hollenstein, K.; Perozo, E.; Locher, K.P. Asymmetry in the structure of the ABC transporter-binding protein complex BtuCD-BtuF. Science 2007, 317, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Borths, E.L.; Locher, K.P.; Lee, A.T.; Rees, D.C. The structure of Escherichia coli BtuF and binding to its cognate ATP binding cassette transporter. Proc. Natl. Acad. Sci. USA 2002, 99, 16642–16647. [Google Scholar] [CrossRef] [PubMed]
- Bhagi-Damodaran, A.; Petrik, I.D.; Marshall, N.M.; Robinson, H.; Lu, Y. Systematic tuning of heme redox potentials and its effects on O2 reduction rates in a designed oxidase in myoglobin. J. Am. Chem. Soc. 2014, 136, 11882–11885. [Google Scholar] [CrossRef] [PubMed]
- Battye, T.G.; Kontogiannis, L.; Johnson, O.; Powell, H.R.; Leslie, A.G. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Cryst. 2011, D67, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Cryst. 2010, D66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Evans, P. Scaling and assessment of data quality. Acta Cryst. 2006, D62, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997, D53, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Cryst. 2010, D66, 486–501. [Google Scholar]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Cryst. 2012, D68, 352–367. [Google Scholar]
- Kitatsuji, C.; Ogura, M.; Uchida, T.; Ishimori, K.; Aono, S. Molecular mechanism for heme-mediated inhibition of 5-aminolevulinic acid synthase 1. Bull. Chem. Soc. Jpn. 2014, 87, 997–1004. [Google Scholar] [CrossRef]
- Okamoto, Y.; Sawai, H.; Ogura, M.; Uchida, T.; Ishimori, K.; Hayashi, T.; Aono, S. Heme-binding properties of HupD functioning as a substrate-binding protein in a heme-uptake ABC-transporter system in Listeria monocytogenes. Bull. Chem. Soc. Jpn. 2014, 87, 1140–1146. [Google Scholar] [CrossRef]
- Sekine, Y.; Tanzawa, T.; Tanaka, Y.; Ishimori, K.; Uchida, T. Cytoplasmic heme-binding protein (HutX) from Vibrio cholera is an intracellular heme transport protein for the heme-degrading enzyme, HutZ. Biochemistry 2016, 55, 884–893. [Google Scholar] [CrossRef] [PubMed]




| CgHmuT Mutants | H141A | Y240A | R242A |
|---|---|---|---|
| Data Collection | |||
| Space group | P41212 | P41212 | P41212 |
| Cell dimensions | |||
| a, b (Å) | 73.38 | 73.01 | 73.06 |
| c (Å) | 147.42 | 145.96 | 147.07 |
| α, β, γ (°) | 90.00 | 90.00 | 90.00 |
| Wavelength (Å) | 0.9000 | 0.9000 | 0.9000 |
| Resolution (Å) | 48.94–1.30 (1.34–1.30) 1 | 35.41–1.65 (1.74–1.65) 1 | 42.27–1.30 (1.34–1.30) 1 |
| Observed Reflection | 935723 | 1416608 | 889463 |
| Unique Reflection | 97636 | 48329 | 96893 |
| Rmerge 2 | 0.058 (0.551) 1 | 0.093 (0.508) 1 | 0.069 (0.487) 1 |
| Rmeas | 0.062 (0.584) 1 | 0.097 (0.527) 1 | 0.073 (0.517) 1 |
| I/σI | 19.98 (3.73) 1 | 16.60 (5.2) 1 | 17.52 (4.58) 1 |
| Completeness (%) | 98.8 (96.6) 1 | 100 (100) 1 | 98.3 (98.3) 1 |
| Refinement | |||
| Resolution (Å) | 48.94–1.30 | 35.41–1.65 | 42.27–1.30 |
| Rwork/Rfree (%) | 15.6/17.3 | 17.6/20.7 | 16.3/18.1 |
| No. of atoms | |||
| Protein | 2314 | 2403 | 2413 |
| Heme | 43 | 86 | 43 |
| Water | 230 | 141 | 225 |
| Average B factors | 14.8 | 23.3 | 16.5 |
| Root-mean-square deviations | |||
| Bond lengths (Å) | 0.005 | 0.015 | 0.007 |
| Bond angles (°) | 1.153 | 1.391 | 1.352 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muraki, N.; Kitatsuji, C.; Ogura, M.; Uchida, T.; Ishimori, K.; Aono, S. Structural Characterization of Heme Environmental Mutants of CgHmuT that Shuttles Heme Molecules to Heme Transporters. Int. J. Mol. Sci. 2016, 17, 829. https://doi.org/10.3390/ijms17060829
Muraki N, Kitatsuji C, Ogura M, Uchida T, Ishimori K, Aono S. Structural Characterization of Heme Environmental Mutants of CgHmuT that Shuttles Heme Molecules to Heme Transporters. International Journal of Molecular Sciences. 2016; 17(6):829. https://doi.org/10.3390/ijms17060829
Chicago/Turabian StyleMuraki, Norifumi, Chihiro Kitatsuji, Mariko Ogura, Takeshi Uchida, Koichiro Ishimori, and Shigetoshi Aono. 2016. "Structural Characterization of Heme Environmental Mutants of CgHmuT that Shuttles Heme Molecules to Heme Transporters" International Journal of Molecular Sciences 17, no. 6: 829. https://doi.org/10.3390/ijms17060829