Arabidopsis Myrosinase Genes AtTGG4 and AtTGG5 Are Root-Tip Specific and Contribute to Auxin Biosynthesis and Root-Growth Regulation
Abstract
:1. Introduction
2. Results
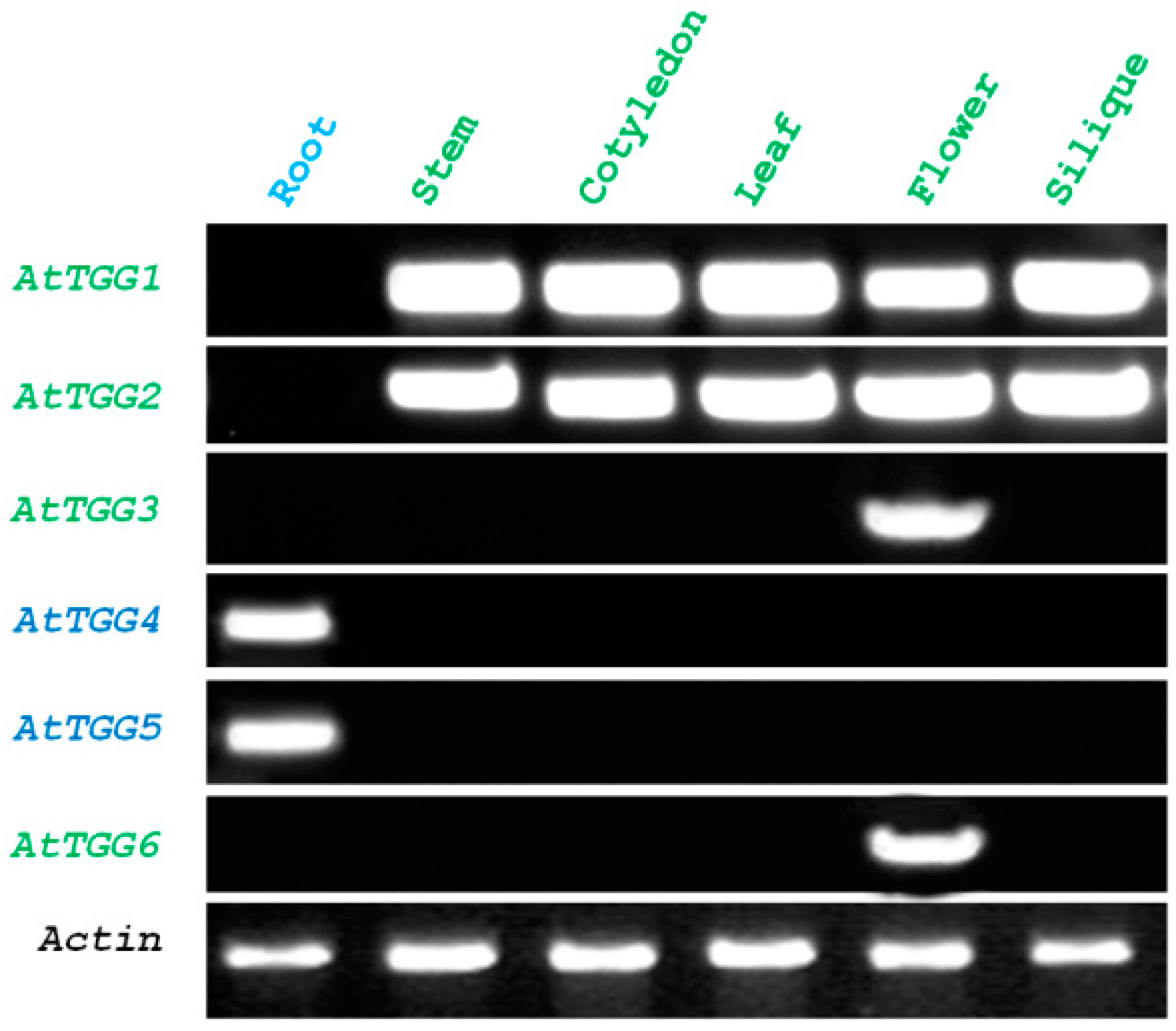
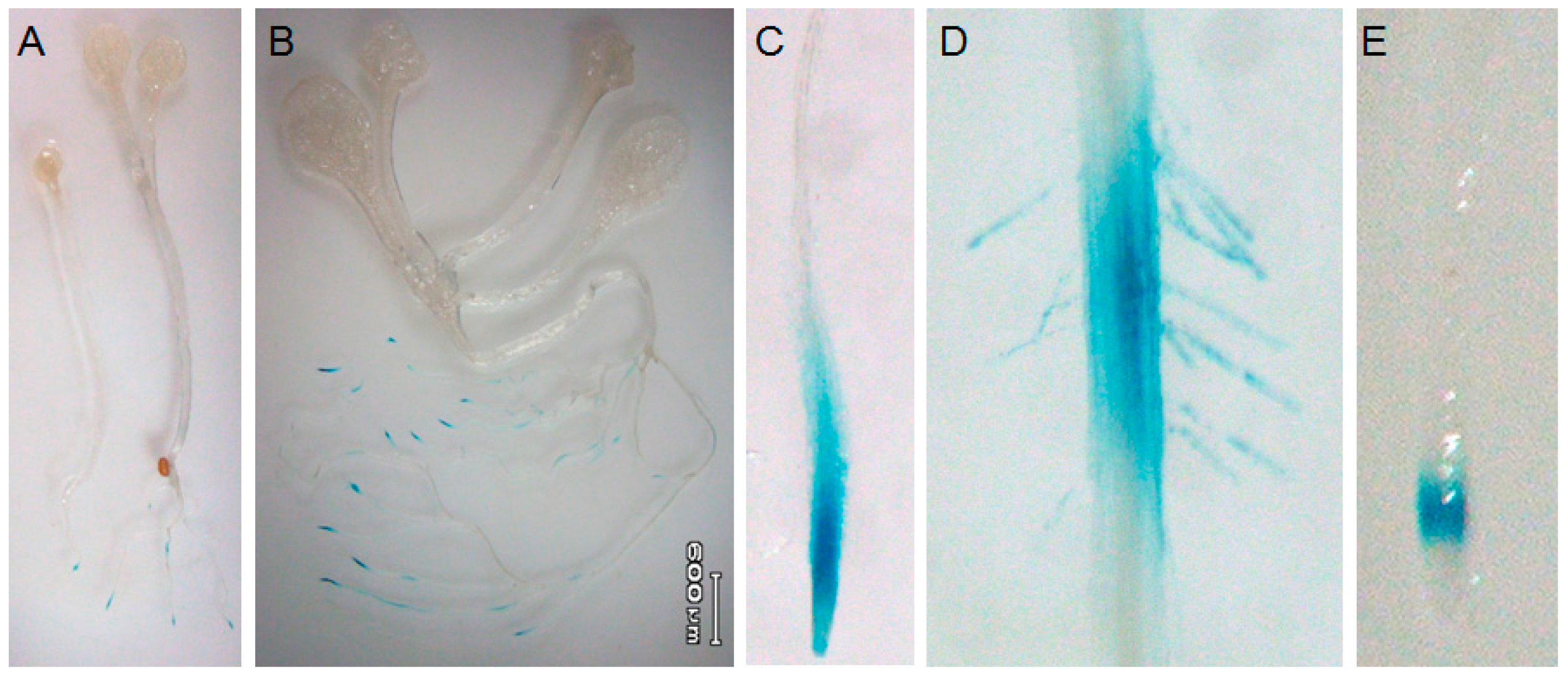
2.1. Root-Tip Specific Expression of AtTGG4 and AtTGG5
2.2. Screening of Homozygous Double T-DNA Insertion Mutants and Myrosinase Activity Test
2.3. AtTGG4 and AtTGG5 Regulate Root Growth
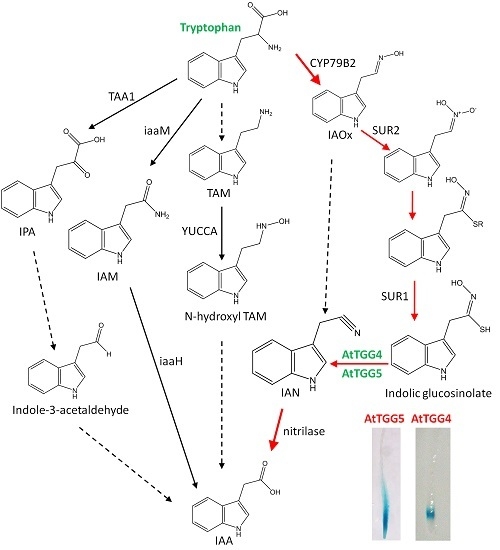
2.4. AtTGG4 and AtTGG5 Contribute to Auxin Biosynthesis in Roots
2.5. AtTGG4 and AtTGG5 Confer Flood-Stress Tolerance in Arabidopsis
3. Discussion
3.1. Root-Tip Specific Expression Implicates a Role of AtTGG4 and AtTGG5 in Root Growth Regulation
3.2. Screening of Homozygous Double T-DNA Insertion Mutants
3.3. AtTGG4 and AtTGG5 Regulate Root Growth and Confer Flood Tolerance
3.4. AtTGG4 and AtTGG5 Contribute to Auxin Biosynthesis through a Tryptophan-Dependent Pathway
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Expression Analysis of Myrosinase Gene Family by Reverse Transcription Polymerase Chain Reaction (RT-PCR)
4.3. Expression Analysis of AtTGG4 Prom::GUS and AtTGG5 Prom::GUS
4.4. Confirmation of T-DNA Insertion Mutants and Creation of Homozygous Double Mutant tgg4tgg5
4.5. Analysis of Myrosinase Activity
4.6. Flood Tolerance Test
4.7. The Role of AtTGG4 and AtTGG5 in Auxin Biosynthesis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rask, L.; Andréasson, E.; Ekbom, B.; Eriksson, S.; Pontoppidan, B.; Meijer, J. Myrosinase: Gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 2000, 42, 93–113. [Google Scholar] [CrossRef] [PubMed]
- Bones, A.M.; Rossiter, J.T. The enzymic and chemically induced decomposition of glucosinolates. Phytochemistry 2006, 67, 1053–1067. [Google Scholar] [CrossRef] [PubMed]
- Raybould, A.F.; Moyes, C.L. The ecological genetics of aliphatic glucosinolates. Heredity 2001, 87, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Fahey, J.W.; Zalcmann, A.T.; Talalay, P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 2001, 56, 5–51. [Google Scholar] [CrossRef]
- Reichelt, M.; Brown, P.D.; Schneider, B.; Oldham, N.J.; Stauber, E.; Tokuhisa, J.; Kliebenstein, D.J.; Mitchell-Olds, T.; Gershenzon, J. Benzoic acid glucosinolate esters and other glucosinolates from Arabidopsis thaliana. Phytochemistry 2002, 59, 663–671. [Google Scholar] [CrossRef]
- Brown, P.D.; Tokuhisa, J.G.; Reichelt, M.; Gershenzon, J. Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 2003, 62, 471–481. [Google Scholar] [CrossRef]
- Petersen, B.L.; Chen, S.; Hansen, C.H.; Olsen, C.E.; Halkier, B.A. Composition and content of glucosinolates in developing Arabidopsis thaliana. Planta 2002, 214, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Brader, G.; Mikkelsen, M.D.; Halkier, B.A.; Tapio Palva, E. Altering glucosinolate profiles modulates disease resistance in plants. Plant J. 2006, 46, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Mewis, I.; Tokuhisa, J.G.; Schultz, J.C.; Appel, H.M.; Ulrichs, C.; Gershenzon, J. Gene expression and glucosinolate accumulation in Arabidopsis thaliana in response to generalist and specialist herbivores of different feeding guilds and the role of defense signaling pathways. Phytochemistry 2006, 67, 2450–2462. [Google Scholar] [CrossRef] [PubMed]
- Bones, A.M.; Rossiter, J.T. The myrosinase-glucosinolate system, its organisation and biochemistry. Physiol. Plant. 1996, 97, 194–208. [Google Scholar] [CrossRef]
- Wang, M.; Li, D.; Sun, X.; Zhu, Y.J.; Nong, H.; Zhang, J. Characterization of a root-specific β-thioglucoside glucohydrolase gene in Carica papaya and its recombinant protein expressed in Pichia pastoris. Plant Sci. 2009, 177, 716–723. [Google Scholar] [CrossRef]
- Xue, J.P.; Lenman, M.; Falk, A.; Rask, L. The glucosinolate-degrading enzyme myrosinase in Brassicaceae is encoded by a gene family. Plant Mol. Biol. 1992, 18, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Falk, A.; Ek, B.; Rask, L. Characterization of a new myrosinase in Brassica napus. Plant Mol. Biol. 1995, 27, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Chadchawan, S.; Bishop, J.; Thangstad, O.P.; Bones, A.M.; Mitchell-Olds, T.; Bradley, D. Arabidopsis cDNA sequence encoding myrosinase. Plant Physiol. 1993, 103, 671–672. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Jorgensen, M.; Pihlgren, U.; Rask, L. The myrosinase gene family in Arabidopsis thaliana: Gene organization, expression and evolution. Plant Mol. Biol. 1995, 27, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Nong, H.; Zhang, J.; Li, D.; Wang, M.; Sun, X.; Zhu, Y.J.; Meijer, J.; Wang, H. Characterization of a novel β-thioglucosidase CpTGG1 in Carica papaya and its substrate-dependent and ascorbic acid-independent O-β-glucosidase activity. J. Integr. Plant Biol. 2010, 52, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Husebye, H.; Chadchawan, S.; Winge, P.; Thangstad, O.P.; Bones, A.M. Guard cell- and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (myrosinase) in Arabidopsis. Plant Physiol. 2002, 128, 1180–1188. [Google Scholar] [CrossRef] [PubMed]
- Andreasson, E.; Bolt Jorgensen, L.; Hoglund, A.S.; Rask, L.; Meijer, J. Different myrosinase and idioblast distribution in Arabidopsis and Brassica napus. Plant Physiol. 2001, 127, 1750–1763. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Pihlgren, U.; Rask, L. Temporal, cell-specific, and tissue-preferential expression of myrosinase genes during embryo and seedling development in Sinapis alba. Planta 1993, 191, 95–101. [Google Scholar] [CrossRef]
- Thangstad, O.P.; Evjen, K.; Bones, A. Immunogold-EM localization of myrosinase in Brassicaceae. Protoplasma 1991, 161, 85–93. [Google Scholar] [CrossRef]
- Kelly, P.J.; Bones, A.; Rossiter, J.T. Sub-cellular immunolocalization of the glucosinolate sinigrin in seedlings of Brassica juncea. Planta 1998, 206, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Koroleva, O.A.; Davies, A.; Deeken, R.; Thorpe, M.R.; Tomos, A.D.; Hedrich, R. Identification of a new glucosinolate-rich cell type in Arabidopsis flower stalk. Plant Physiol. 2000, 124, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Andersson, D.; Meijer, J. Characterisation of a new type of beta-thioglucoside glycosidase (myrosinase) in Arabidopsis thaliana. In Proceedings of the VIII International Conference on Arabidopsis Research, Sevilla, Spain, 28 June–2 July 2002; pp. 6–25.
- Andersson, D.; Chakrabarty, R.; Bejai, S.; Zhang, J.; Rask, L.; Meijer, J. Myrosinases from root and leaves of Arabidopsis thaliana have different catalytic properties. Phytochemistry 2009, 70, 1345–1354. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, D.; Tan, D.; Gong, S.; Sun, X.; Meijer, J.; Zhang, J. The two non-functional myrosinase genes TGG3 and TGG6 in Arabidopsis are expressed predominantly in pollen. Plant Sci. 2009, 177, 371–375. [Google Scholar] [CrossRef]
- Zhang, J.; Pontoppidan, B.; Xue, J.; Rask, L.; Meijer, J. The third myrosinase gene TGG3 in Arabidopsis thaliana is a pseudogene specifically expressed in stamen and petal. Physiol. Plant. 2002, 115, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Han, B.; Tan, D.; Wang, M.; Ding, M.; Zhang, J. Identification and evolution of functional alleles of the previously described pollen specific myrosinase pseudogene AtTGG6 in Arabidopsis thaliana. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Rask, L. The unusual 5′ splicing border GC is used in myrosinase genes of the Brassicaceae. Plant Mol. Biol. 1995, 29, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Olafsdottir, E.S.; Bolt Jorgensen, L.; Jaroszewski, J.W. Cyanogenesis in glucosinolate-producing plants: Carica papaya and Carica quercifolia. Phytochemistry 2002, 60, 269–273. [Google Scholar] [CrossRef]
- Cairns, T.; Siegmund, E.G.; Stamp, J.J.; Jacobs, R.M. Characterization of benzyl isothiocyanate and phenyl acetonitrile from papayas by mass spectrometry. J. Assoc. Off. Anal. Chem. 1988, 71, 547–550. [Google Scholar] [PubMed]
- Flath, R.A.; Forrey, R.R. Volatile components of papaya (Carica papaya L., Solo variety). J. Agric. Food Chem. 1977, 25, 103–109. [Google Scholar] [CrossRef]
- Ulmasov, T.; Murfett, J.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997, 9, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Barth, C.; Jander, G. Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 2006, 46, 549–562. [Google Scholar] [CrossRef] [PubMed]
- Lukowitz, W.; Gillmor, C.S.; Scheible, W.R. Positional cloning in Arabidopsis: Why it feels good to have a genome initiative working for you. Plant Physiol. 2000, 123, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Bednarek, P.; Pislewska-Bednarek, M.; Svatos, A.; Schneider, B.; Doubsky, J.; Mansurova, M.; Humphry, M.; Consonni, C.; Panstruga, R.; Sanchez-Vallet, A.; et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 2009, 323, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Nitz, I.; Berkefeld, H.; Puzio, P.S.; Grundler, F.M. Pyk10, a seedling and root specific gene and promoter from Arabidopsis thaliana. Plant Sci. 2001, 161, 337–346. [Google Scholar] [CrossRef]
- Dan, H.; Yang, G.; Zheng, Z.L. A negative regulatory role for auxin in sulphate deficiency response in Arabidopsis thaliana. Plant Mol. Biol. 2007, 63, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, W.P.; Cottaz, S.; Driguez, H.; Iori, R.; Palmieri, S.; Henrissat, B. The crystal structures of Sinapis alba myrosinase and a covalent glycosyl-enzyme intermediate provide insights into the substrate recognition and active-site machinery of an S-glycosidase. Structure 1997, 5, 663–675. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Sairam, R.K.; Kumutha, D.; Ezhilmathi, K.; Deshmukh, P.S.; Srivastava, G.C. Physiology and biochemistry of waterlogging tolerance in plants. Biol. Plant. 2008, 52, 401–412. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hou, X.; Tsuge, T.; Ding, M.; Aoyama, T.; Oka, A.; Gu, H.; Zhao, Y.; Qu, L.J. The possible action mechanisms of indole-3-acetic acid methyl ester in Arabidopsis. Plant Cell Rep. 2008, 27, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, R.; Ma, C.J.; Vlot, A.C.; Klessig, D.F.; Pichersky, E. Inactive methyl indole-3-acetic acid ester can be hydrolyzed and activated by several esterases belonging to the AtMES esterase family of Arabidopsis. Plant Physiol. 2008, 147, 1034–1045. [Google Scholar] [CrossRef] [PubMed]
- Seidel, C.; Walz, A.; Park, S.; Cohen, J.D.; Ludwig-Muller, J. Indole-3-acetic acid protein conjugates: Novel players in auxin homeostasis. Plant Biol. 2006, 8, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Walz, A.; Seidel, C.; Rusak, G.; Park, S.; Cohen, J.D.; Ludwig-Muller, J. Heterologous expression of IAP1, a seed protein from bean modified by indole-3-acetic acid, in Arabidopsis thaliana and Medicago truncatula. Planta 2008, 227, 1047–1061. [Google Scholar] [CrossRef] [PubMed]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef] [PubMed]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef] [PubMed]
- Kutachek, M.; Prochazka, Z.; Veres, K. Biogenesis of glucobrassicin, the in vitro precursor of ascorbigen. Nature 1962, 194, 393–394. [Google Scholar] [CrossRef]
- Underhill, E.W.; Wetter, L.R.; Chisholm, M.D. Biosynthesis of glucosinolates. Biochem. Soc. Symp. 1973, 303–326. [Google Scholar]
- Du, L.; Lykkesfeldt, J.; Olsen, C.E.; Halkier, B.A. Involvement of cytochrome P450 in oxime production in glucosinolate biosynthesis as demonstrated by an in vitro microsomal enzyme system isolated from jasmonic acid-induced seedlings of Sinapis alba L. Proc. Natl. Acad. Sci. USA 1995, 92, 12505–12509. [Google Scholar] [CrossRef] [PubMed]
- Hull, A.K.; Vij, R.; Celenza, J.L. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 2379–2384. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hull, A.K.; Gupta, N.R.; Goss, K.A.; Alonso, J.; Ecker, J.R.; Normanly, J.; Chory, J.; Celenza, J.L. Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Gene Dev. 2002, 16, 3100–3112. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, M.D.; Naur, P.; Halkier, B.A. Arabidopsis mutants in the C–S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 2004, 37, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Boerjan, W.; Cervera, M.T.; Delarue, M.; Beeckman, T.; Dewitte, W.; Bellini, C.; Caboche, M.; van Onckelen, H.; van Montagu, M.; Inze, D. Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 1995, 7, 1405–1419. [Google Scholar] [CrossRef] [PubMed]
- An, Y.Q.; McDowell, J.M.; Huang, S.; McKinney, E.C.; Chambliss, S.; Meagher, R.B. Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 1996, 10, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Bechtold, N.; Ellis, J.; Pelletier, G. In planta agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 1993, 316, 1194–1199. [Google Scholar]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. Gus fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [PubMed]







| Gametes | TGG4-tgg5E12 (≈44%) | tgg4E8-TGG5 (≈44%) | TGG4-TGG5 (≈6%) | tgg4E8-tgg5E12 (≈6%) |
|---|---|---|---|---|
| TGG4-tgg5E12 (≈44%) | TGG4-tgg5E12 | tgg4E8-TGG5 | TGG4-TGG5 | tgg4E8-tgg5E12 |
| TGG4-tgg5E12 | TGG4-tgg5E12 | TGG4-tgg5E12 | TGG4-tgg5E12 | |
| (≈19.4%) | (≈19.4%) | (≈2.6%) | (≈2.6%) | |
| tgg4E8-TGG5 (≈44%) | TGG4-tgg5E12 | tgg4E8-TGG5 | TGG4-TGG5 | tgg4E8-tgg5E12 |
| tgg4E8-TGG5 | tgg4E8-TGG5 | tgg4E8-TGG5 | tgg4E8-TGG5 | |
| (≈19.4%) | (≈19.4%) | (≈2.6%) | (≈2.6%) | |
| TGG4-TGG5 (≈6%) | TGG4-tgg5E12 | tgg4E8-TGG5 | TGG4-TGG5 | tgg4E8-tgg5E12 |
| TGG4-TGG5 | TGG4-TGG5 | TGG4-TGG5 | TGG4-TGG5 | |
| (≈2.6%) | (≈2.6%) | (≈0.36%) | (≈0.36%) | |
| tgg4E8-tgg5E12 (≈6%) | TGG4-tgg5E12 | tgg4E8-TGG5 | TGG4-TGG5 | tgg4E8-tgg5E12 |
| tgg4E8-tgg5E12 | tgg4E8-tgg5E12 | tgg4E8-tgg5E12 | tgg4E8-tgg5E12 | |
| (≈2.6%) | (≈2.6%) | (≈0.36%) | (≈0.36%) |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, L.; Wang, M.; Han, B.; Tan, D.; Sun, X.; Zhang, J. Arabidopsis Myrosinase Genes AtTGG4 and AtTGG5 Are Root-Tip Specific and Contribute to Auxin Biosynthesis and Root-Growth Regulation. Int. J. Mol. Sci. 2016, 17, 892. https://doi.org/10.3390/ijms17060892
Fu L, Wang M, Han B, Tan D, Sun X, Zhang J. Arabidopsis Myrosinase Genes AtTGG4 and AtTGG5 Are Root-Tip Specific and Contribute to Auxin Biosynthesis and Root-Growth Regulation. International Journal of Molecular Sciences. 2016; 17(6):892. https://doi.org/10.3390/ijms17060892
Chicago/Turabian StyleFu, Lili, Meng Wang, Bingying Han, Deguan Tan, Xuepiao Sun, and Jiaming Zhang. 2016. "Arabidopsis Myrosinase Genes AtTGG4 and AtTGG5 Are Root-Tip Specific and Contribute to Auxin Biosynthesis and Root-Growth Regulation" International Journal of Molecular Sciences 17, no. 6: 892. https://doi.org/10.3390/ijms17060892
APA StyleFu, L., Wang, M., Han, B., Tan, D., Sun, X., & Zhang, J. (2016). Arabidopsis Myrosinase Genes AtTGG4 and AtTGG5 Are Root-Tip Specific and Contribute to Auxin Biosynthesis and Root-Growth Regulation. International Journal of Molecular Sciences, 17(6), 892. https://doi.org/10.3390/ijms17060892