Evidence of Decoupling Protein Structure from Spidroin Expression in Spider Dragline Silks
Abstract
:1. Introduction
2. Results and Discussion
2.1. Amino Acid Compositions
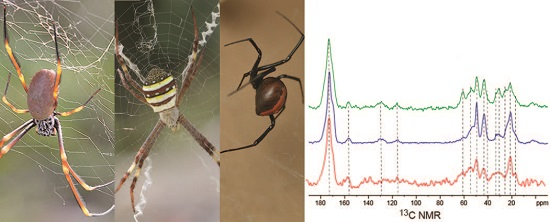
2.2. Solid-State Nuclear Magnetic Resonance Spectroscopy
2.3. Influences Affecting Mechanical Properties
3. Experimental Section
3.1. Spiders and Silk Collecting
3.2. High Performance Liquid Chromatography (HPLC)
3.3. Solid-State NMR (ssNMR) Spectroscopy
3.4. Tensile Testing
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Heim, M.; Keerl, D.; Scheibel, T. Spider silk: From soluble protein to extraordinary fiber. Angew. Chem. Int. Ed. 2009, 48, 3584–3596. [Google Scholar] [CrossRef] [PubMed]
- Van Krevelen, D.W. Properties of Polymers: Their Correlation with Chemical Structure, Their Numerical Estimation and Predictions from Additive Group Contributions; Elsevier: Amsterdam, The Netherlands, 2009. [Google Scholar]
- Parkhe, A.D.; Seeley, S.K.; Gardner, A.; Thompson, L.; Lewis, R.V. Structural studies of spider silk proteins in the fiber. J. Mol. Recog. 1997, 10, 1–6. [Google Scholar] [CrossRef]
- Erdogan, H.; Apaydin, M.S. Incorporating amino acid typing into nuclear magnetic resonance protein structure-based assignments. Proteom. Bioinform. 2012, 5, 116–121. [Google Scholar] [CrossRef]
- Fratzl, P.; Jacob, H.F.; Rimmerthaler, S.; Roschger, P.; Klaushofer, K. Position resolved small angle X-ray scattering of complex biological materials. J. Appl. Crystal. 1997, 30, 765–769. [Google Scholar] [CrossRef]
- Ene, R.; Papadopoulos, P.; Kramer, F. Combined structural model of spider dragline silk. Soft Matter 2009, 5, 4568–4574. [Google Scholar] [CrossRef]
- Rathore, O.; Sogah, D.Y. Nanostructure formation through β-sheet self-assembly in silk-based materials. Macromolecules 2001, 34, 1477–1486. [Google Scholar] [CrossRef]
- Bovey, F.A.; Miran, P.A. NMR of Polymers; Academic Press: San Diego, CA, USA, 1996. [Google Scholar]
- An, B.; Hinman, M.; Holland, G.P.; Yarger, J.L.; Lewis, R.V. Inducing β-Sheets formation in synthetic spider silk fibers by aqueous post-spin stretching. Biomacromolecules 2011, 12, 2375–2381. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Yarger, J.L.; Holland, G.P. Elucidating proline dynamics in spider dragline silk fibre using 2H–13C HETCOR MAS NMR. Chem. Commun. 2014, 50, 4856–4859. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liivak, O.; Seidel, A.; LaVerde, G.; Zax, D.B.; Jelinski, L.W. Supercontraction and backbone dynamics in spider silk: 13C and 2H NMR Studies. J. Am. Chem. Soc. 2000, 122, 9019–9025. [Google Scholar] [CrossRef]
- Li, X.; Eles, P.T.; Michal, C.A. Water permeability of spider dragline silk. Biomacromolecules 2009, 10, 1270–1275. [Google Scholar] [CrossRef] [PubMed]
- Creager, M.S.; Jenkins, J.E.; Thagard-Yeaman, L.A.; Brooks, A.E.; Jones, J.A.; Lewis, R.V.; Holland, G.P.; Yarger, J.L. Solid-state NMR comparison of various spiders' dragline silk fiber. Biomacromolecules 2010, 11, 2039–2043. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, J.E.; Creager, M.S.; Lewis, R.V.; Holland, G.P.; Yarger, J.L. Quantitative correlation between the protein primary sequences and secondary structures in spider dragline silks. Biomacromolecules 2010, 11, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Yarger, J.L.; Holland, G.P. Exploring the backbone dynamics of native spider silk proteins in black widow silk glands with solution-state NMR spectroscopy. Polymer 2014, 55, 3879–3885. [Google Scholar] [CrossRef]
- Shi, X.; Holland, G.P.; Yarger, J.L. Molecular dynamics of spider dragline silk fiber by 2H MAS NMR. Biomacromolecules 2015, 16, 852–859. [Google Scholar] [CrossRef] [PubMed]
- Izdebski, T.; Akhenblit, P.; Jenkins, J.E.; Yarger, J.L.; Holland, G.P. Structure and dynamics of aromatic residues in spider silk: 2D carbon correlation NMR of dragline fibers. Biomacromolecules 2010, 11, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Rauscher, S.; Baud, S.; Miao, M.; Keeley, F.W.; Pomes, R. Proline and glycine control protein self-organization into elastomeric or amyloid fibrils. Structure 2006, 14, 1667–1676. [Google Scholar] [CrossRef] [PubMed]
- Holland, G.P.; Yarger, J.L.; Lewis, R.V. WISE NMR characterization of nanoscale heterogeneity and mobility in supercontracted Nephila clavipes spider dragline silk. J. Am. Chem. Soc. 2004, 126, 5867–5872. [Google Scholar] [CrossRef] [PubMed]
- Addison, J.B.; Ashton, N.N.; Weber, W.S.; Stewart, R.J.; Holland, G.P.; Yarger, J.L. β-Sheet nanocrystalline domains formed from phosphorylated serine-rich motifs in caddisfly larval silk: A solid state NMR and XRD study. Biomacromolecues 2013, 11, 1140–1148. [Google Scholar] [CrossRef] [PubMed]
- Holland, G.P.; Jenkins, J.E.; Creager, M.S.; Lewis, R.V.; Yarger, J.L. Solid-state NMR investigation of major and minor ampullate spider silk in the native and hydrated states. Biomacromolecules 2008, 9, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Shi, X.; Thompson, F.; Weber, W.S.; Mou, Q.; Yarger, J.L. Protein secondary structure of Green Lynx spider dragline silk investigated by solid-state NMR and X-ray diffraction. Int. J. Biol. Macromol. 2015, 81, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, J.E.; Sampath, S.; Butler, E.; Kim, J.; Henning, R.W.; Holland, G.P.; Yarger, J.L. Characterizing the secondary protein structure of black widow dragline silk using solid-state NMR and X-ray diffraction. Biomacromolecules 2013, 14, 3472–3483. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Johnson, M.A.; Martin, D.C. Microstructural characterization of Bombyx mori silk fibers. Macromolecules 1998, 31, 8857–8864. [Google Scholar] [CrossRef]
- Asakura, T.; Yang, M.; Kawase, T.; Nakazawa, Y. 13C solid-state NMR Study of structural heterogeneity in peptides containing both polyalanine and repeated GGA sequences as a local structural model of Nephila clavipes dragline silk (Spidroin 1). Macromolecules 2005, 38, 3356–3363. [Google Scholar] [CrossRef]
- Craig, C.L.; Riekel, C. Comparative architecture of silks, fibrous proteins and their encoding genes in insects and spiders. Comp. Biochem. Physiol. B 2002, 133, 493–507. [Google Scholar] [CrossRef]
- Savage, K.N.; Gosline, J.M. The role of proline in the elastic mechanism of hydrated spider silks. J. Exp. Biol. 2008, 211, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shao, Z.Z.; Vollrath, F. Elasticity of spider silks. Biomacromolecules 2008, 9, 1782–1786. [Google Scholar] [CrossRef] [PubMed]
- Boutry, C.; Blackledge, T.A. Evolution of supercontraction in spider silk: Structure–function relationship from tarantulas to orb-weavers. J. Exp. Biol. 2010, 213, 3505–3514. [Google Scholar] [CrossRef] [PubMed]
- Van Beek, J.D.; Hess, S.; Vollrath, F.; Meier, B.H. The molecular structure of spider dragline silk: Folding and orientation of the protein backbone. Proc. Nat. Acad. Sci. USA 2002, 99, 10266–10271. [Google Scholar] [CrossRef] [PubMed]
- Blamires, S.J.; Wu, C.L.; Tso, I.M. Variation in protein intake induces variation in spider silk expression. PLoS ONE 2012, 7, e31626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blamires, S.J.; Wu, C.L.; Blackledge, T.A.; Tso, I.M. Environmentally induced post-spin property changes in spider silks: Influences of web type, spidroin composition and ecology. Biol. J. Linn. Soc. 2012, 106, 580–588. [Google Scholar] [CrossRef]
- Blamires, S.J.; Wu, C.L.; Blackledge, T.A.; Tso, I.M. Post-secretion processing influences spider silk performance. J. R. Soc. Interface 2012, 9, 2479–2487. [Google Scholar] [CrossRef] [PubMed]
- Craig, C.L.; Hsu, M.; Kaplan, D.L.; Pierce, M.E. A comparison of the composition of silk proteins produced by spiders and insects. Int. J. Biol. Macromol. 1999, 24, 109–118. [Google Scholar] [CrossRef]
- Guehrs, K.H.; Schlott, B.; Grosse, F.; Weisshart, K. Environmental conditions impinge on dragline silk prtotein composition. Insect Mol. Biol. 2008, 17, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Craig, C.L.; Riekel, C.; Herberstein, M.E.; Weber, R.S.; Kaplan, D.; Pierce, N.E. Evidence for diet effects on the composition of silk proteins produced by spiders. Mol. Biol. Evol. 2000, 17, 1904–1913. [Google Scholar] [CrossRef] [PubMed]
- Tso, I.M.; Wu, H.C.; Hwang, I.R. Giant wood spider Nephila pilipes alters silk protein in response to prey variation. J. Exp. Biol. 2005, 208, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- Blamires, S.J.; Liao, C.P.; Chang, C.K.; Chuang, Y.C.; Wu, C.L.; Blackledge, T.A.; Sheu, H.S.; Tso, I.M. Mechanical performance of spider silk is robust to nutrient-mediated changes in protein composition. Biomacromolecules 2015, 16, 1218–1225. [Google Scholar] [CrossRef] [PubMed]
- Marhabaie, M.; Leeper, T.C.; Blackledge, T.A. Protein composition correlates with the mechanical properties of spider (Argiope trifasciata) dragline silk. Biomacromolecules 2014, 15, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.P.; Chi, K.; Tso, I.M. The effects of wind on trap structural and material properties of a sit-and-wait predator. Behav. Ecol. 2009, 20, 1194–1203. [Google Scholar] [CrossRef]
- Dicko, C.; Vollrath, F.; Kennedy, J.M. Spider silk protein folding is controlled by changing pH. Biomacromolecules 2004, 5, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Hedhammer, M.; Rising, A.; Grip, S.; Saenz Martinez, A.; Nordling, K.; Casals, C.; Stark, M.; Johansson, J. Structural properties of recombinant nonrepetitive and repetitive parts of major ampullate spidroin 1 from Euprosthenops australis: Implications for fiber formation. Biochemistry 2008, 47, 3407–3417. [Google Scholar] [CrossRef] [PubMed]
- Knight, D.P.; Knight, M.M.; Vollrath, F. Beta transitions and stress-induced phase separation in the spinning of spider dragline silk. Int. J. Biol. Macromol. 2000, 27, 205–210. [Google Scholar] [CrossRef]
- Vollrath, F.; Knight, D.P. Liquid crystalline spinning of spider silk. Nature 2001, 410, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Lefevre, T.; Paquet-Mercier, F.; Rioux-Dube, J.F.; Pezolet, M. Structure of silk by Raman spectromicroscopy: From the spinning glands to the fibers. Biopolymers 2011, 97, 322–335. [Google Scholar] [CrossRef] [PubMed]
- Giesa, T.; Perry, C.C.; Buehler, M.J. Secondary structure and critical stress for a model of spider silk assembly. Biomacromolecules 2016, 17, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, N.A.; Garb, J.E.; Tinghitella, R.M.; Collin, M.A.; Hayashi, C.Y. Blueprint for a high-performance biomaterial: Full-length spider dragline silk genes. PLoS ONE 2007, 2, e514. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, A.C.; Sima, Y.H.; Lu, C.; Xiang, X.H.; Nakagaki, M. The molecular structures of major ampullate silk proteins of the wasp spider, Argiope bruennichi: A second blueprint for synthesizing de novo silk. Comp. Biochem. Physiol. B 2013, 164, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Work, R.W.; Young, T.C. The amino acid compositions of major ampullate and minor ampullate silks of certain orb-web building spiders (Araneae, Araneidae). J. Arachnol. 1987, 15, 65–80. [Google Scholar]
- Chaw, R.C.; Correa-Garhwal, S.M.; Clarke, T.H.; Ayoub, N.A.; Hayashi, C.Y. Proteomic evidence for components of spider silk synthesis from black widow silk glands and fibers. J. Proteome Res. 2015, 14, 4223–4231. [Google Scholar] [CrossRef] [PubMed]
- Elices, M.; Plaza, G.R.; Perz-Riguero, J.; Guinea, G.V. The hidden link between supercontraction and mechanical behaviour of spider silks. J. Mech. Behav. Biomed. Mater. 2011, 4, 658–669. [Google Scholar] [CrossRef] [PubMed]
- Gosline, J.M.; Denny, M.W.; De Monte, M.E. Spider silk as rubber. Nature 1984, 309, 551–552. [Google Scholar] [CrossRef]
- Vollrath, F. Strength and structure of spider’s silk. Rev. Mol. Biotechnol. 2000, 74, 67–83. [Google Scholar] [CrossRef]
- Perz-Riguero, J.; Elices, M.; Guinea, G.V. Controlled supercontraction tailors the tensile behaviour of spider silk. Polymer 2003, 44, 3733–3736. [Google Scholar] [CrossRef]
- Grubb, D.T.; Jelinski, L.W. Fiber morphology of spider silk: The effects of tensile deformation. Macromolecules 1997, 30, 2860–2867. [Google Scholar] [CrossRef]
- Lewis, R.V. Bionanotechnology: Proteins to Nanodevices; Renugopalakrishnan, V., Lewis, R.V., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 61–78. [Google Scholar]
- Meyers, M.A.; Chawla, K.K. Mechanical Behavior of Materials; Cambridge University Press: Cambridge, UK, 2009. [Google Scholar]


| Amino Acids (Percentage Composition) | |||||
|---|---|---|---|---|---|
| Glutamine | Serine | Glycine | Alanine | Proline | |
| (a) May | |||||
| Argiope keyserlingi | 7.90 (0.88) | 7.02 (0.72) | 35.39 (1.52) | 26.46 (1.87) | 4.17 (0.84) |
| Nephila plumipes | 6.39 (0.92) | 4.77 (0.33) | 41.09 (0.45) | 30.23 (0.56) | 2.71 (0.42) |
| Latrodectus hasselti | 9.61 (0.20) | 6.78 (0.62) | 37.56 (1.67) | 26.0 (1.92) | 3.13 (0.65) |
| (b) November | |||||
| Argiope keyserlingi | 8.27 (0.84) | 7.34 (2.29) | 34.61 (6.77) | 19.34 (6.76) * | 12.53 (1.67) * |
| Latrodectus hasselti | 8.99 (0.38) | 7.47 (0.46) | 32.28 (2.75) | 29.55 (2.92) | 2.79 (0.65) |
| (c) Sequenced compositions | |||||
| Latrodectus hesperus MaSp1 | 6.9 | – | 33.5 | 31.1 | 0.4 |
| Latrodectus hesperus MaSp2 | 11.3 | – | 42.3 | 32.7 | 8.6 |
| Argiope bruennichi MaSp1 | 4.38 | 5.67 | 45.05 | 30.61 | 0 |
| Argiope bruennichi MaSp2 | 14.50 | 3.04 | 38.47 | 22.51 | 12.48 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blamires, S.J.; Kasumovic, M.M.; Tso, I.-M.; Martens, P.J.; Hook, J.M.; Rawal, A. Evidence of Decoupling Protein Structure from Spidroin Expression in Spider Dragline Silks. Int. J. Mol. Sci. 2016, 17, 1294. https://doi.org/10.3390/ijms17081294
Blamires SJ, Kasumovic MM, Tso I-M, Martens PJ, Hook JM, Rawal A. Evidence of Decoupling Protein Structure from Spidroin Expression in Spider Dragline Silks. International Journal of Molecular Sciences. 2016; 17(8):1294. https://doi.org/10.3390/ijms17081294
Chicago/Turabian StyleBlamires, Sean J., Michael M. Kasumovic, I-Min Tso, Penny J. Martens, James M. Hook, and Aditya Rawal. 2016. "Evidence of Decoupling Protein Structure from Spidroin Expression in Spider Dragline Silks" International Journal of Molecular Sciences 17, no. 8: 1294. https://doi.org/10.3390/ijms17081294