Analysis of Small RNAs in Streptococcus mutans under Acid Stress—A New Insight for Caries Research
Abstract
:1. Introduction
2. Results
2.1. Growth and pH Drop Assay Results with an Initial pH of 5.5
2.2. Sequence Analysis and Identification of sRNAs
2.3. Experimental Validation of Predicted sRNAs and Prediction of Target mRNA
2.4. Bacterial Growth and Vitality Assessment under Different Acid Stress Conditions
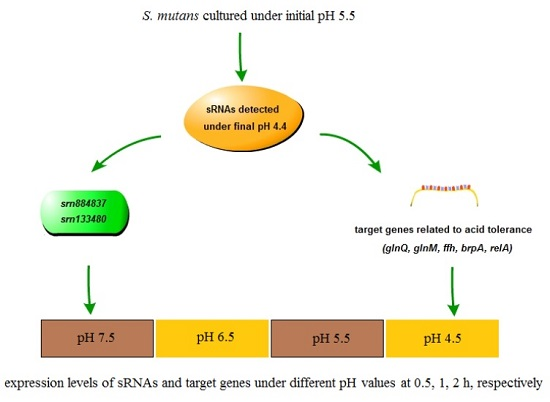
2.5. Expression Analysis of sRNAs and Target mRNAs under Different Acid Stress Conditions
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Total RNA Isolation
4.3. Small RNA Library Construction and Deep Sequencing
4.4. Bioinformatics Analysis of Sequence Data and Identification of sRNAs
4.5. Validation of sRNA Candidates and Prediction of Verified sRNA Targets
4.6. Effect of Different Acid Stress Conditions on the Growth, Vitality, and Gene Expression of S. mutans
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ramos-Jorge, J.; Pordeus, I.A.; Ramos-Jorge, M.L.; Marques, L.S.; Paiva, S.M. Impact of untreated dental caries on quality of life of preschool children: Different stages and activity. Community Dent. Oral Epidemiol. 2014, 42, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Jorge, J.; Alencar, B.M.; Pordeus, I.A.; Soares, M.E.; Marques, L.S.; Ramos-Jorge, M.L.; Paiva, S.M. Impact of dental caries on quality of life among preschool children: Emphasis on the type of tooth and stages of progression. Eur. J. Oral Sci. 2015, 123, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Martins-Junior, P.A.; Vieira-Andrade, R.G.; Correa-Faria, P.; Oliveira-Ferreira, F.; Marques, L.S.; Ramos-Jorge, M.L. Impact of early childhood caries on the oral health-related quality of life of preschool children and their parents. Caries Res. 2013, 47, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.Q. Report of the Third National Oral Health Survey in China; People’s Medical Publishing House: Beijing, China, 2008. (In Chinese) [Google Scholar]
- Kang, K.H.; Lee, J.S.; Yoo, M.; Jin, I. The influence of HtrA expression on the growth of Streptococcus mutans during acid stress. Mol. Cells 2010, 29, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Matsui, R.; Cvitkovitch, D. Acid tolerance mechanisms utilized by Streptococcus mutans. Future Microbiol. 2010, 5, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Neilands, J.; Sutherland, D.; Resin, A.; Wejse, P.L.; Chavez de Paz, L.E. Chitosan nanoparticles affect the acid tolerance response in adhered cells of Streptococcus mutans. Caries Res. 2011, 45, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Lemos, J.A.; Luzardo, Y.; Burne, R.A. Physiologic effects of forced down-regulation of dnaK and groEL expression in Streptococcus mutans. J. Bacteriol. 2007, 189, 1582–1588. [Google Scholar] [CrossRef] [PubMed]
- Crowley, P.J.; Svensater, G.; Snoep, J.L.; Bleiweis, A.S.; Brady, L.J. An ffh mutant of Streptococcus mutans is viable and able to physiologically adapt to low pH in continuous culture. FEMS Microbiol. Lett. 2004, 234, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Hanna, M.N.; Ferguson, R.J.; Li, Y.H.; Cvitkovitch, D.G. uvrA is an acid-inducible gene involved in the adaptive response to low pH in Streptococcus mutans. J. Bacteriol. 2001, 183, 5964–5973. [Google Scholar] [CrossRef] [PubMed]
- Lemos, J.A.; Chen, Y.Y.; Burne, R.A. Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J. Bacteriol. 2001, 183, 6074–6084. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.T.; Baker, H.V.; Burne, R.A. Influence of BrpA on critical virulence attributes of Streptococcus mutans. J. Bacteriol. 2006, 188, 2983–2992. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, S.; Matsumoto-Nakano, M.; Fujita, K.; Nagayama, K.; Funao, J.; Ooshima, T. Effects of recombinase A deficiency on biofilm formation by Streptococcus mutans. Oral Microbiol. Immunol. 2009, 24, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Krastel, K.; Senadheera, D.B.; Mair, R.; Downey, J.S.; Goodman, S.D.; Cvitkovitch, D.G. Characterization of a glutamate transporter operon, glnQHMP, in Streptococcus mutans and its role in acid tolerance. J. Bacteriol. 2010, 192, 984–993. [Google Scholar] [CrossRef] [PubMed]
- Lemos, J.A.; Brown, T.A., Jr.; Burne, R.A. Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect. Immun. 2004, 72, 1431–1440. [Google Scholar] [CrossRef] [PubMed]
- Caldelari, I.; Chao, Y.; Romby, P.; Vogel, J. RNA-mediated regulation in pathogenic bacteria. Cold Spring Harb. Perspect. Med. 2013, 3, a010298. [Google Scholar] [CrossRef] [PubMed]
- Hoe, C.H.; Raabe, C.A.; Rozhdestvensky, T.S.; Tang, T.H. Bacterial sRNAs: Regulation in stress. Int. J. Med. Microbiol. 2013, 303, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Lalaouna, D.; Simoneau-Roy, M.; Lafontaine, D.; Masse, E. Regulatory RNAs and target mRNA decay in prokaryotes. Biochim. Biophys. Acta 2013, 1829, 742–747. [Google Scholar] [CrossRef] [PubMed]
- Geissmann, T.; Chevalier, C.; Cros, M.J.; Boisset, S.; Fechter, P.; Noirot, C.; Schrenzel, J.; Francois, P.; Vandenesch, F.; Gaspin, C.; et al. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 2009, 37, 7239–7257. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Xia, W.; Li, S.; Li, W.; Liu, J.; Ding, H.; Li, J.; Li, H.; Chen, Y.; Su, X.; et al. Identification and expression of small non-coding RNA, L10-Leader, in different growth phases of Streptococcus mutans. Nucleic Acid Ther. 2012, 22, 177–186. [Google Scholar] [PubMed]
- Chabelskaya, S.; Gaillot, O.; Felden, B. A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule. PLoS Pathog. 2010, 6, e1000927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izar, B.; Mraheil, M.A.; Hain, T. Identification and role of regulatory non-coding RNAs in Listeria monocytogenes. Int. J. Mol. Sci. 2011, 12, 5070–5079. [Google Scholar] [CrossRef] [PubMed]
- Khandige, S.; Kronborg, T.; Uhlin, B.E.; Moller-Jensen, J. sRNA-Mediated Regulation of P-Fimbriae Phase Variation in Uropathogenic Escherichia coli. PLoS Pathog. 2015, 11, e1005109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mika, F.; Hengge, R. Small Regulatory RNAs in the Control of Motility and Biofilm Formation in E. coli and Salmonella. Int. J. Mol. Sci. 2013, 14, 4560–4579. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P.; Ross, H.F.; Projan, S.J.; Kornblum, J.; Kreiswirth, B.; Moghazeh, S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993, 12, 3967–3975. [Google Scholar] [PubMed]
- Pellin, D.; Miotto, P.; Ambrosi, A.; Cirillo, D.M.; Di Serio, C. A genome-wide identification analysis of small regulatory RNAs in Mycobacterium tuberculosis by RNA-Seq and conservation analysis. PLoS ONE 2012, 7, e32723. [Google Scholar] [CrossRef] [PubMed]
- Schiano, C.A.; Koo, J.T.; Schipma, M.J.; Caulfield, A.J.; Jafari, N.; Lathem, W.W. Genome-wide analysis of small RNAs expressed by Yersinia pestis identifies a regulator of the Yop-Ysc type III secretion system. J. Bacteriol. 2014, 196, 1659–1670. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Arya, S.; Patil, S.D.; Sharma, A.; Jain, P.K.; Navani, N.K.; Pathania, R. Identification of novel regulatory small RNAs in Acinetobacter baumannii. PLoS ONE 2014, 9, e93833. [Google Scholar] [CrossRef] [PubMed]
- Warrier, I.; Hicks, L.D.; Battisti, J.M.; Raghavan, R.; Minnick, M.F. Identification of novel small RNAs and characterization of the 6S RNA of Coxiella burnetii. PLoS ONE 2014, 9, e100147. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Xu, X.; Li, X.; Liu, S.; Lei, S.; Yang, M.; Yu, J.; Yuan, J.; Ke, Y.; Du, X.; et al. Large-scale identification of small noncoding RNA with strand-specific deep sequencing and characterization of a novel virulence-related sRNA in Brucella melitensis. Sci. Rep. 2016, 6, 25123. [Google Scholar] [CrossRef] [PubMed]
- Acebo, P.; Martin-Galiano, A.J.; Navarro, S.; Zaballos, A.; Amblar, M. Identification of 88 regulatory small RNAs in the TIGR4 strain of the human pathogen Streptococcus pneumoniae. RNA 2012, 18, 530–546. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Hong, S.H. Analysis of microRNA-size, small RNAs in Streptococcus mutans by deep sequencing. FEMS Microbiol. Lett. 2012, 326, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.Y.; Yang, Y.M.; Li, K.Z.; Lei, L.; Li, M.; Yang, Y.; Tao, X.; Yin, J.X.; Zhang, R.; Ma, X.R.; et al. The rnc Gene Promotes Exopolysaccharide Synthesis and Represses the vicRKX Gene Expressions via MicroRNA-Size Small RNAs in Streptococcus mutans. Front. Microbiol. 2016, 7, 687. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, J.; Gunasekaran, P. Computational small RNA prediction in bacteria. Bioinform. Biol. Insights 2013, 7, 83–95. [Google Scholar] [PubMed]
- Yi, S.; Gao, Z.X.; Zhao, H.; Zeng, C.; Luo, W.; Chen, B.; Wang, W.M. Identification and characterization of microRNAs involved in growth of blunt snout bream (Megalobrama amblycephala) by Solexa sequencing. BMC Genom. 2013, 14, 754. [Google Scholar] [CrossRef] [PubMed]
- Downey, J.S.; Mashburn-Warren, L.; Ayala, E.A.; Senadheera, D.B.; Hendrickson, W.K.; McCall, L.W.; Sweet, J.G.; Cvitkovitch, D.G.; Spatafora, G.A.; Goodman, S.D. In vitro manganese-dependent cross-talk between Streptococcus mutans VicK and GcrR: Implications for overlapping stress response pathways. PLoS ONE 2014, 9, e115975. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.S.; Park, S.J.; Kim, M.K.; Kim, Y.H.; Lee, S.B.; Lee, K.H.; Choi, N.Y.; Lee, Y.R.; Lee, Y.E.; You, Y.O. Inhibitory effects of chrysanthemum boreale essential oil on biofilm formation and virulence factor expression of Streptococcus mutans. Evid. Based Complement. Altern. Med. 2015, 2015, 616309. [Google Scholar] [CrossRef] [PubMed]
- Len, A.C.; Harty, D.W.; Jacques, N.A. Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology 2004, 150 Pt 5, 1339–1351. [Google Scholar] [CrossRef] [PubMed]
- Diez-Martinez, R.; Garcia-Fernandez, E.; Manzano, M.; Martinez, A.; Domenech, M.; Vallet-Regi, M.; Garcia, P. Auranofin-loaded nanoparticles as a new therapeutic tool to fight streptococcal infections. Sci. Rep. 2016, 6, 19525. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Lei, R.; Henninger, T.D.; Contreras, L.M. Discovery of ethanol-responsive small RNAs in Zymomonas mobilis. Appl. Environ. Microbiol. 2014, 80, 4189–4198. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.M.; Darfeuille, F.; Plantinga, T.H.; Vogel, J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007, 21, 2804–2817. [Google Scholar] [CrossRef] [PubMed]
- Pain, A.; Ott, A.; Amine, H.; Rochat, T.; Bouloc, P.; Gautheret, D. An assessment of bacterial small RNA target prediction programs. RNA Biol. 2015, 12, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Norden-Krichmar, T.M.; Allen, A.E.; Gaasterland, T.; Hildebrand, M. Characterization of the small RNA transcriptome of the diatom, Thalassiosira pseudonana. PLoS ONE 2011, 6, e22870. [Google Scholar] [CrossRef] [PubMed]
- Vacca Smith, A.M.; Ng-Evans, L.; Wunder, D.; Bowen, W.H. Studies concerning the glucosyltransferase of Streptococcus sanguis. Caries Res. 2000, 34, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]






| ID | Start Location | Length | Sequence | Strands | Mismatch | Counts (Average) | Ct (Average) |
|---|---|---|---|---|---|---|---|
| srn884837 | 1930112 | 29 | ACGTGAATCATCGGTGCCAATACAGCATT | + | 0 | 1143 | 28.47 |
| srn133480 | 293197 | 27 | TAAGCGATGTAAGCTGTGTGCTCTATT | + | 0 | 2238 | 32.83 |
| srn854592 | 1864362 | 21 | AAAAAGAGCAGCTAAATCGGA | − | 0 | 949 | 33.38 |
| srn140177 | 305931 | 18 | TTTGTTCAAGATTGTACT | + | 0 | 170 | 35.67 |
| srn470015 | 991733 | 19 | TCTAAGACAAATTCCGTTA | − | 0 | 114 | 36.27 |
| srn371778 | 763825 | 18 | TAACATCTGAAACTAAGG | + | 0 | 185 | 36.58 |
| srn821712 | 1793874 | 18 | TTGACTGACTAACTATCA | + | 0 | 35 | 36.90 |
| srn228002 | 480025 | 20 | TAGTATCTGTAGTTGCTGCA | + | 0 | 299 | 37.09 |
| srn638035 | 1357678 | 19 | TGTCTCAGTCCTATACACA | − | 0 | 8 | 37.30 |
| srn219672 | 461702 | 20 | GATCAATACATGTATCCTTA | − | 0 | 48 | 38.38 |
| srn91608 | 210391 | 18 | AAGTGTCTAAGTTAGATT | − | 0 | 139 | - |
| srn174875 | 374257 | 18 | GGACAGGATGTCTACTTA | − | 0 | 101 | - |
| srn342303 | 713515 | 18 | GGACAGTATCTTCAATTA | − | 0 | 177 | - |
| srn430462 | 899609 | 18 | AGAGTATTTAACTAGTCG | − | 0 | 69 | - |
| srn444332 | 929474 | 20 | ACCAAACGATCAAACCGTGA | − | 0 | 352 | - |
| srn628738 | 1328165 | 35 | ACACAGCTCTAAAACTCACCATATTAATTAATGGC | − | 0 | 1451 | - |
| srn800380 | 1757893 | 18 | ATTAAGACCCCCAACAAT | − | 0 | 436 | - |
| srn821539 | 1793724 | 19 | TTGTTTTAGAAACTTCTGC | + | 0 | 44 | - |
| ID | Primer Sequences |
|---|---|
| srn884837 | purchased from RiboBio, Guangzhou, China |
| srn133480 | purchased from RiboBio, Guangzhou, China |
| glnQ (F) | GACAGGTTGTTGTTTTACTTG |
| glnQ (R) | GGTCCTTAGTTGAAGCATTGG |
| glnM (F) | GAAGTCATTCGCTCTGGTATTGAAG |
| glnM (R) | CATTGGTGGCAAGATAGTTCTGATG |
| ffh (F) | AAGGTAAGCAAGTCTCCCATTC |
| ffh (R) | TCCGTCAAATCACTGGAAAAC |
| brpA (F) | GGAGGAGCTGCATCAGGATTC |
| brpA (R) | AACTCCAGCACATCCAGCAAG |
| relA (F) | ACAAAAAGGGTATCGTCCGTACAT |
| relA (R) | AATCACGCTTGGTATTGCTAATTG |
| 16S rRNA (F) | CTGACTTGAGTGCAGAAGGGGA |
| 16S rRNA (R) | CGTCAGTGACAGACCAGAGAGC |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Tao, Y.; Yu, L.; Zhuang, P.; Zhi, Q.; Zhou, Y.; Lin, H. Analysis of Small RNAs in Streptococcus mutans under Acid Stress—A New Insight for Caries Research. Int. J. Mol. Sci. 2016, 17, 1529. https://doi.org/10.3390/ijms17091529
Liu S, Tao Y, Yu L, Zhuang P, Zhi Q, Zhou Y, Lin H. Analysis of Small RNAs in Streptococcus mutans under Acid Stress—A New Insight for Caries Research. International Journal of Molecular Sciences. 2016; 17(9):1529. https://doi.org/10.3390/ijms17091529
Chicago/Turabian StyleLiu, Shanshan, Ye Tao, Lixia Yu, Peilin Zhuang, Qinghui Zhi, Yan Zhou, and Huancai Lin. 2016. "Analysis of Small RNAs in Streptococcus mutans under Acid Stress—A New Insight for Caries Research" International Journal of Molecular Sciences 17, no. 9: 1529. https://doi.org/10.3390/ijms17091529
APA StyleLiu, S., Tao, Y., Yu, L., Zhuang, P., Zhi, Q., Zhou, Y., & Lin, H. (2016). Analysis of Small RNAs in Streptococcus mutans under Acid Stress—A New Insight for Caries Research. International Journal of Molecular Sciences, 17(9), 1529. https://doi.org/10.3390/ijms17091529