Reverse Genetics Approaches for the Development of Influenza Vaccines
Abstract
:1. Influenza Virus
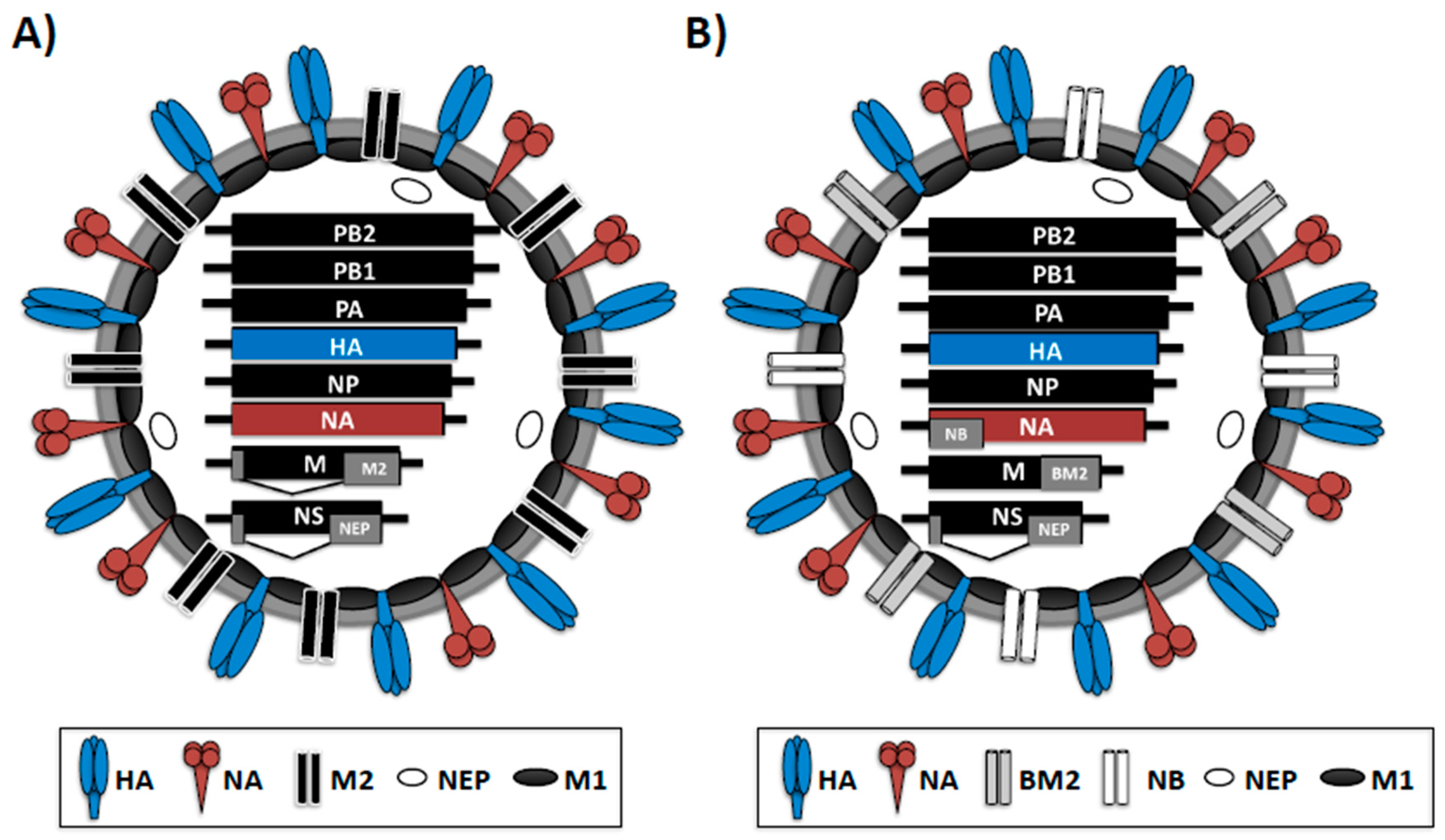
1.1. Influenza Virus Structure and Genome Organization
1.2. Influenza Life Cycle
1.3. Influenza Viruses and Their Impact on Human Health
1.4. Current Strategies to Combat Human Influenza Infections
1.4.1. Influenza Vaccines
1.4.2. Influenza Antivirals
2. Influenza Vaccine Production
2.1. Vaccine Strain Selection
2.2. Types of Influenza Seasonal Vaccines
2.2.1. Influenza Inactivated Vaccine (IIV)
2.2.2. Live-Attenuated Influenza Vaccine
2.3. Substrates for the Production of Influenza Vaccines
2.3.1. Chicken Embryonated Eggs
2.3.2. Cell Cultures
2.4. Adjuvants
3. Influenza Reverse Genetics for Vaccine Development
3.1. Influenza Reverse Genetics
3.2. Reverse Genetics for the Investigation of Influenza Virus
3.3. Generation of Recombinant Influenza Viruses Using Reverse Genetics Approaches
3.4. Influenza Reverse Genetics for the Development of IIV
3.5. Influenza Reverse Genetics Approaches for the Development of Live-Attenuated Influenza Vaccines (LAIVs)
3.6. Taking Advantage of Reverse Genetics for the Development of Novel Vaccines
3.6.1. NS1 Truncated or Deficient Viruses as LAIVs
3.6.2. Codon-Deoptimized LAIVs
3.6.3. Single-Cycle Infectious Viruses as LAIVs
3.6.4. LAIVs Based on the Rearrangement of the Influenza Viral Genome
3.6.5. Other Approaches to Generate Influenza Vaccines
4. Conclusions and Future Direction
Acknowledgments
Conflicts of Interest
Abbreviations
| IAV | Influenza A virus |
| IBV | Influenza B virus |
| vRNA | Viral RNAs |
| vRNP | Viral ribonucleoprotein |
| NAb | Neutralizing antibody |
| ts | Temperature-sensitive |
| ca | Cold-adapted |
| att | Attenuated |
| PB2 | Polymerase basic 2 |
| PB1 | Polymerase basic 1 |
| PA | Polymerase acid |
| HA | Hemagglutinin |
| NA | Neuraminidase |
| NP | Nucleoprotein |
| NS1 | Non-structural protein 1 |
| ORF | Open reading frame |
| NCR | Non-coding region |
| IFN-I | Type I interferon |
| LAIV | Live attenuated influenza vaccine |
| IIV | Influenza inactivated vaccine |
| VLP | Virus-like particle |
| WHO | World Health Organization |
References
- Palese, P.S.; Shaw, M.L. Orthomyxoviridae: The viruses and their replication. In Fields Virology, 5th ed.; Knipe, D.M., Howley, P.M., Griffin, D.E., Lamb, R.A., Martin, M.A., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007. [Google Scholar]
- Steel, J.; Lowen, A.C.; Mubareka, S.; Palese, P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 2009, 5, e1000252. [Google Scholar] [CrossRef] [PubMed]
- Parrish, C.R.; Kawaoka, Y. The origins of new pandemic viruses: The acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu. Rev. Microbiol. 2005, 59, 553–586. [Google Scholar] [CrossRef] [PubMed]
- Bean, W.J.; Schell, M.; Katz, J.; Kawaoka, Y.; Naeve, C.; Gorman, O.; Webster, R.G. Evolution of the H3 influenza virus hemagglutinin from human and nonhuman hosts. J. Virol. 1992, 66, 1129–1138. [Google Scholar] [PubMed]
- Chen, R.; Holmes, E.C. The evolutionary dynamics of human influenza B virus. J. Mol. Evol. 2008, 66, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Osterhaus, A.D.; Rimmelzwaan, G.F.; Martina, B.E.; Bestebroer, T.M.; Fouchier, R.A. Influenza B virus in seals. Science 2000, 288, 1051–1053. [Google Scholar] [CrossRef] [PubMed]
- Resa-Infante, P.; Jorba, N.; Coloma, R.; Ortin, J. The influenza virus rna synthesis machine: Advances in its structure and function. RNA Biol. 2011, 8, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Brownlee, G.G.; Fodor, E.; Kawaoka, Y. Orthomyxovirus replication, transcription, and polyadenylation. Curr. Top. Microbiol. Immunol. 2004, 283, 121–143. [Google Scholar] [PubMed]
- Huang, T.S.; Palese, P.; Krystal, M. Determination 0of influenza virus proteins required for genome replication. J. Virol. 1990, 64, 5669–5673. [Google Scholar] [PubMed]
- Wright, P.; Nuemann, G.; Kawaoka, Y. Orthomhxoviruses. In Fields Virology, 5th ed.; Knipe, D.M., Howley, P.M., Griffin, D.E., Lamb, R.A., Martin, M.A., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007. [Google Scholar]
- Stoeckle, M.Y.; Shaw, M.W.; Choppin, P.W. Segment-specific and common nucleotide sequences in the noncoding regions of influenza B virus genome RNAs. Proc. Natl. Acad. Sci. USA 1987, 84, 2703–2707. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.F.; Nogales, A.; Finch, C.; Tuffy, K.M.; Domm, W.; Perez, D.R.; Topham, D.J.; Martinez-Sobrido, L. Influenza A and B virus intertypic reassortment through compatible viral packaging signals. J. Virol. 2014, 88, 10778–10791. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.; Elderfield, R.A.; Barclay, W.S. Molecular studies of influenza B virus in the reverse genetics era. J. Gen. Virol. 2011, 92, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Wise, H.M.; Hutchinson, E.C.; Jagger, B.W.; Stuart, A.D.; Kang, Z.H.; Robb, N.; Schwartzman, L.M.; Kash, J.C.; Fodor, E.; Firth, A.E.; et al. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog. 2012, 8, e1002998. [Google Scholar] [CrossRef] [PubMed]
- Horvath, C.M.; Williams, M.A.; Lamb, R.A. Eukaryotic coupled tanslation of tandem cistrons: Identification of the influenza B virus BM2 polypeptide. EMBO J. 1990, 9, 2639–2647. [Google Scholar] [PubMed]
- Skehel, J.J.; Wiley, D.C. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef] [PubMed]
- Varghese, J.N.; McKimm-Breschkin, J.L.; Caldwell, J.B.; Kortt, A.A.; Colman, P.M. The structure of the complex between influenza virus neuraminidase and sialic acid, the viral receptor. Proteins 1992, 14, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Tulip, W.R.; Varghese, J.N.; Laver, W.G.; Webster, R.G.; Colman, P.M. Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex. J. Mol. Biol. 1992, 227, 122–148. [Google Scholar] [CrossRef]
- Yoon, S.W.; Webby, R.J.; Webster, R.G. Evolution and ecology of influenza A viruses. Curr. Top. Microbiol. Immunol. 2014, 385, 359–375. [Google Scholar] [PubMed]
- Tong, S.; Zhu, X.; Li, Y.; Shi, M.; Zhang, J.; Bourgeois, M.; Yang, H.; Chen, X.; Recuenco, S.; Gomez, J.; et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013, 9, e1003657. [Google Scholar] [CrossRef] [PubMed]
- McCullers, J.A.; Saito, T.; Iverson, A.R. Multiple genotypes of influenza B virus circulated between 1979 and 2003. J. Virol. 2004, 78, 12817–12828. [Google Scholar] [CrossRef] [PubMed]
- Kanegae, Y.; Sugita, S.; Endo, A.; Ishida, M.; Senya, S.; Osako, K.; Nerome, K.; Oya, A. Evolutionary pattern of the hemagglutinin gene of influenza B viruses isolated in japan: Cocirculating lineages in the same epidemic season. J. Virol. 1990, 64, 2860–2865. [Google Scholar] [PubMed]
- Thompson, W.W.; Shay, D.K.; Weintraub, E.; Brammer, L.; Cox, N.; Anderson, L.J.; Fukuda, K. Mortality associated with influenza and respiratory syncytial virus in the united states. JAMA 2003, 289, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Li, W.C.; Shih, S.R.; Huang, Y.C.; Chen, G.W.; Chang, S.C.; Hsiao, M.J.; Tsao, K.C.; Lin, T.Y. Clinical and genetic characterization of severe influenza B-associated diseases during an outbreak in Taiwan. J. Clin. Virol. 2008, 42, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Hite, L.K.; Glezen, W.P.; Demmler, G.J.; Munoz, F.M. Medically attended pediatric influenza during the resurgence of the victoria lineage of influenza B virus. Int. J. Infect. Dis. 2007, 11, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Pinto, L.H.; Holsinger, L.J.; Lamb, R.A. Influenza virus M2 protein has ion channel activity. Cell 1992, 69, 517–528. [Google Scholar] [CrossRef]
- Lamb, R.A.; Choppin, P.W. The gene structure and replication of influenza virus. Annu. Rev. Biochem. 1983, 52, 467–506. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, E.C.; von Kirchbach, J.C.; Gog, J.R.; Digard, P. Genome packaging in influenza A virus. J. Gen. Virol. 2010, 91, 313–328. [Google Scholar] [CrossRef] [PubMed]
- Rossman, J.S.; Lamb, R.A. Influenza virus assembly and budding. Virology 2011, 411, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Gerber, M.; Isel, C.; Moules, V.; Marquet, R. Selective packaging of the influenza A genome and consequences for genetic reassortment. Trends Microbiol. 2014, 22, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Hale, B.G.; Randall, R.E.; Ortin, J.; Jackson, D. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 2008, 89, 2359–2376. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.; Fodor, E. Emerging roles for the influenza A virus nuclear export protein (NEP). PLoS Pathog. 2012, 8, e1003019. [Google Scholar] [CrossRef] [PubMed]
- Ampofo, W.K.; Baylor, N.; Cobey, S.; Cox, N.J.; Daves, S.; Edwards, S.; Ferguson, N.; Grohmann, G.; Hay, A.; Katz, J.; et al. Improving influenza vaccine virus selection: Report of a WHO informal consultation held at WHO headquarters, geneva, switzerland, 14–16 June 2010. Influenza Other Respir. Viruses 2013, 7, 52–53. [Google Scholar] [PubMed]
- Barr, I.G.; McCauley, J.; Cox, N.; Daniels, R.; Engelhardt, O.G.; Fukuda, K.; Grohmann, G.; Hay, A.; Kelso, A.; Klimov, A.; et al. Epidemiological, antigenic and genetic characteristics of seasonal influenza A (H1N1), a(H3N2) and B influenza viruses: Basis for the WHO recommendation on the composition of influenza vaccines for use in the 2009–2010 northern hemisphere season. Vaccine 2010, 28, 1156–1167. [Google Scholar] [CrossRef] [PubMed]
- Cox, N.J.; Brammer, T.L.; Regnery, H.L. Influenza: Global surveillance for epidemic and pandemic variants. Eur. J. Epidemiol. 1994, 10, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.M.; Song, J.M.; Compans, R.W. Novel vaccines against influenza viruses. Virus Res. 2011, 162, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P., 3rd; Walsh, E.E. Influenza: Evolving strategies in treatment and prevention. Semin. Respir. Crit. Care Med. 2007, 28, 144–158. [Google Scholar] [CrossRef] [PubMed]
- Molinari, N.A.; Ortega-Sanchez, I.R.; Messonnier, M.L.; Thompson, W.W.; Wortley, P.M.; Weintraub, E.; Bridges, C.B. The annual impact of seasonal influenza in the us: Measuring disease burden and costs. Vaccine 2007, 25, 5086–5096. [Google Scholar] [CrossRef] [PubMed]
- Girard, M.P.; Cherian, T.; Pervikov, Y.; Kieny, M.P. A review of vaccine research and development: Human acute respiratory infections. Vaccine 2005, 23, 5708–5724. [Google Scholar] [CrossRef] [PubMed]
- Keech, M.; Beardsworth, P. The impact of influenza on working days lost: A review of the literature. Pharmacoeconomics 2008, 26, 911–924. [Google Scholar] [CrossRef] [PubMed]
- Paul Glezen, W.; Schmier, J.K.; Kuehn, C.M.; Ryan, K.J.; Oxford, J. The burden of influenza B: A structured literature review. Am. J. Public Health 2013, 103, e43–e51. [Google Scholar] [CrossRef] [PubMed]
- Gasparini, R.; Amicizia, D.; Lai, P.L.; Panatto, D. Clinical and socioeconomic impact of seasonal and pandemic influenza in adults and the elderly. Hum. Vaccines Immunother. 2012, 8, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.J.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef] [PubMed]
- Kilbourne, E.D. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 2006, 12, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Carrat, F.; Flahault, A. Influenza vaccine: The challenge of antigenic drift. Vaccine 2007, 25, 6852–6862. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lindstrom, S.E.; Shaw, M.W.; Smith, C.B.; Hall, H.E.; Mungall, B.A.; Subbarao, K.; Cox, N.J.; Klimov, A. Reassortment and evolution of current human influenza A and B viruses. Virus Res. 2004, 103, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Sandbulte, M.R.; Westgeest, K.B.; Gao, J.; Xu, X.; Klimov, A.I.; Russell, C.A.; Burke, D.F.; Smith, D.J.; Fouchier, R.A.; Eichelberger, M.C. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proc. Natl. Acad. Sci. USA 2011, 108, 20748–20753. [Google Scholar] [CrossRef] [PubMed]
- Shaw, M.L.; Palese, P. Orthomyxoviridae. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013. [Google Scholar]
- Mikheeva, A.; Ghendon, Y.Z. Intrinsic interference between influenza A and B viruses. Arch. Virol. 1982, 73, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Knipe, D.M.; Howley, P.M. Fields Virology, 6th ed.; Wolters Kluwer/Lippincott Williams & Wilkins Health: Philadelphia, PA, USA, 2013; p. 2. [Google Scholar]
- Taubenberger, J.K.; Morens, D.M. 1918 influenza: The mother of all pandemics. Emerg. Infect. Dis. 2006, 12, 15–22. [Google Scholar] [CrossRef] [PubMed]
- CDC. Update: Infections with a swine—Origin influenza A (H1N1) virus—United States and other countries, April 28, 2009. MMWR Morb. Mortal. Wkly. Rep. 2009, 58, 431–433. [Google Scholar]
- WHO. Pandemic (H1N1) 2009—Update 112. Available online: http://www.who.int/csr/don/2010_08_06/en/index.html (accessed on 20 December 2016).
- Uyeki, T.M.; Cox, N.J. Global concerns regarding novel influenza A (H7N9) virus infections. N. Engl. J. Med. 2013, 368, 1862–1864. [Google Scholar] [CrossRef] [PubMed]
- Yen, H.L.; Webster, R.G. Pandemic influenza as a current threat. Curr. Top. Microbiol. Immunol. 2009, 333, 3–24. [Google Scholar] [PubMed]
- Brotherton, J.M.; Delpech, V.C.; Gilbert, G.L.; Hatzi, S.; Paraskevopoulos, P.D.; McAnulty, J.M. A large outbreak of influenza A and B on a cruise ship causing widespread morbidity. Epidemiol. Infect. 2003, 130, 263–271. [Google Scholar] [CrossRef] [PubMed]
- CDC. From the centers for disease control and prevention. Influenza B virus outbreak in a cruise ship—Northern europe, 2000. JAMA 2001, 285, 1833–1834. [Google Scholar]
- DeStefano, F. An outbreak of influenza B at an indiana boarding school: Estimate of vaccine efficacy. Public Health Rep. 1982, 97, 269–272. [Google Scholar] [PubMed]
- Fayinka, O.A.; Balayan, M.S.; Kirya, G.B.; Rugyendo, W. An outbreak of influenza B in a closed community school in Uganda. East. Afr. Med. J. 1977, 54, 6–8. [Google Scholar] [PubMed]
- Gordon Smith, C.E.; Thomson, W.G. An outbreak of influenza due to type B virus in a residential boys’ school in Malaya. Med. J. Malaya 1956, 10, 332–337. [Google Scholar] [PubMed]
- Osterholm, M.T.; Kelley, N.S.; Sommer, A.; Belongia, E.A. Efficacy and effectiveness of influenza vaccines: A systematic review and meta-analysis. Lancet Infect. Dis. 2012, 12, 36–44. [Google Scholar] [CrossRef]
- De Jong, M.D.; Tran, T.T.; Truong, H.K.; Vo, M.H.; Smith, G.J.; Nguyen, V.C.; Bach, V.C.; Phan, T.Q.; Do, Q.H.; Guan, Y.; et al. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N. Engl. J. Med. 2005, 353, 2667–2672. [Google Scholar] [CrossRef] [PubMed]
- Hurt, A.C.; Holien, J.K.; Parker, M.; Kelso, A.; Barr, I.G. Zanamivir-resistant influenza viruses with a novel neuraminidase mutation. J. Virol. 2009, 83, 10366–10373. [Google Scholar] [CrossRef] [PubMed]
- Monto, A.S.; McKimm-Breschkin, J.L.; Macken, C.; Hampson, A.W.; Hay, A.; Klimov, A.; Tashiro, M.; Webster, R.G.; Aymard, M.; Hayden, F.G.; et al. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob. Agents Chemother. 2006, 50, 2395–2402. [Google Scholar] [CrossRef] [PubMed]
- Belshe, R.B.; Edwards, K.M.; Vesikari, T.; Black, S.V.; Walker, R.E.; Hultquist, M.; Kemble, G.; Connor, E.M. Live attenuated versus inactivated influenza vaccine in infants and young children. N. Engl. J. Med. 2007, 356, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Belshe, R.B.; Newman, F.K.; Wilkins, K.; Graham, I.L.; Babusis, E.; Ewell, M.; Frey, S.E. Comparative immunogenicity of trivalent influenza vaccine administered by intradermal or intramuscular route in healthy adults. Vaccine 2007, 25, 6755–6763. [Google Scholar] [CrossRef] [PubMed]
- Pronker, E.S.; Claassen, E.; Osterhaus, A.D. Development of new generation influenza vaccines: Recipes for success? Vaccine 2012, 30, 7344–7347. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.M.; Patriarca, P.A.; Treanor, J. Flublok, a recombinant hemagglutinin influenza vaccine. Influenza. Other Respir. Viruses 2008, 2, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.F.; Guo, H.; Albrecht, R.A.; Garcia-Sastre, A.; Topham, D.J.; Martinez-Sobrido, L. Protection against lethal influenza with a viral mimic. J. Virol. 2013, 87, 8591–8605. [Google Scholar] [CrossRef] [PubMed]
- Sun, W. Approval Letter—Fluarix Quadrivalent (BL 125127/513); U.S. Food and Drug Administration (FDA): Silver Spring, MD, USA, 2012.
- Belongia, E.A.; Kieke, B.A.; Donahue, J.G.; Greenlee, R.T.; Balish, A.; Foust, A.; Lindstrom, S.; Shay, D.K. Effectiveness of inactivated influenza vaccines varied substantially with antigenic match from the 2004–2005 season to the 2006–2007 season. J. Infect. Dis. 2009, 199, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Pica, N.; Palese, P. Toward a universal influenza virus vaccine: Prospects and challenges. Annu. Rev. Med. 2013, 64, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Gorse, G.J.; Belshe, R.B.; Munn, N.J. Superiority of live-attenuated compared with inactivated influenza A virus vaccines in older, chronically ill adults. Chest 1991, 100, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Sastre, A. Antiviral response in pandemic influenza viruses. Emerg. Infect. Dis. 2006, 12, 44–47. [Google Scholar] [CrossRef] [PubMed]
- Oxford, J.S.; Bossuyt, S.; Balasingam, S.; Mann, A.; Novelli, P.; Lambkin, R. Treatment of epidemic and pandemic influenza with neuraminidase and M2 proton channel inhibitors. Clin. Microbiol. Infect. 2003, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Seibert, C.W.; Kaminski, M.; Philipp, J.; Rubbenstroth, D.; Albrecht, R.A.; Schwalm, F.; Stertz, S.; Medina, R.A.; Kochs, G.; Garcia-Sastre, A.; et al. Oseltamivir-resistant variants of the 2009 pandemic H1N1 influenza A virus are not attenuated in the guinea pig and ferret transmission models. J. Virol. 2010, 84, 11219–11226. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.J.; Cooper, K.L.; Tappenden, P.; Rees, A.; Simpson, E.L.; Read, R.C.; Nicholson, K.G. Oseltamivir, zanamivir and amantadine in the prevention of influenza: A systematic review. J. Infect. 2011, 62, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, J.T.; Hoopes, J.D.; Le, M.H.; Smee, D.F.; Patick, A.K.; Faix, D.J.; Blair, P.J.; de Jong, M.D.; Prichard, M.N.; Went, G.T. Triple combination of amantadine, ribavirin, and oseltamivir is highly active and synergistic against drug resistant influenza virus strains in vitro. PLoS ONE 2010, 5, e9332. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.J.; Wolstenholme, A.J.; Skehel, J.J.; Smith, M.H. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985, 4, 3021–3024. [Google Scholar] [PubMed]
- Gubareva, L.V.; Kaiser, L.; Hayden, F.G. Influenza virus neuraminidase inhibitors. Lancet 2000, 355, 827–835. [Google Scholar] [CrossRef]
- Burnham, A.J.; Baranovich, T.; Govorkova, E.A. Neuraminidase inhibitors for influenza B virus infection: Efficacy and resistance. Antivir. Res. 2013, 100, 520–534. [Google Scholar] [CrossRef] [PubMed]
- Farrukee, R.; Mosse, J.; Hurt, A.C. Review of the clinical effectiveness of the neuraminidase inhibitors against influenza B viruses. Expert Rev. Anti Infect. Ther. 2013, 11, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- Pizzorno, A.; Abed, Y.; Boivin, G. Influenza drug resistance. Semin. Respir. Crit. Care Med. 2011, 32, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Sheu, T.G.; Mishin, V.P.; Klimov, A.I.; Gubareva, L.V. Assessment of pandemic and seasonal influenza A (H1N1) virus susceptibility to neuraminidase inhibitors in three enzyme activity inhibition assays. Antimicrob. Agents Chemother. 2010, 54, 3671–3677. [Google Scholar] [CrossRef] [PubMed]
- Ampofo, W.K.; Al Busaidy, S.; Cox, N.J.; Giovanni, M.; Hay, A.; Huang, S.; Inglis, S.; Katz, J.; Mokhtari-Azad, T.; Peiris, M.; et al. Strengthening the influenza vaccine virus selection and development process. In Presented at the 2nd WHO Informal Consultation for Improving Influenza Vaccine Virus Selection, Geneva, Switzerland, 7–9 December 2011.
- Russell, C.A.; Jones, T.C.; Barr, I.G.; Cox, N.J.; Garten, R.J.; Gregory, V.; Gust, I.D.; Hampson, A.W.; Hay, A.J.; Hurt, A.C.; et al. Influenza vaccine strain selection and recent studies on the global migration of seasonal influenza viruses. Vaccine 2008, 26, D31–D34. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Mozdzanowska, K.; Palladino, G.; Gerhard, W. Heterosubtypic immunity to influenza type A virus in mice. Effector mechanisms and their longevity. J. Immunol. 1994, 152, 1653–1661. [Google Scholar] [PubMed]
- Medina, R.A.; Manicassamy, B.; Stertz, S.; Seibert, C.W.; Hai, R.; Belshe, R.B.; Frey, S.E.; Basler, C.F.; Palese, P.; Garcia-Sastre, A. Pandemic 2009 H1N1 vaccine protects against 1918 Spanish influenza virus. Nat. Commun. 2010, 1, 28. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Palese, P.; Steel, J. Advances in universal influenza virus vaccine design and antibody mediated therapies based on conserved regions of the hemagglutinin. Curr. Top. Microbiol. Immunol. 2015, 386, 301–321. [Google Scholar] [PubMed]
- Milian, E.; Kamen, A.A. Current and emerging cell culture manufacturing technologies for influenza vaccines. BioMed Res. Int. 2015, 2015, 504831. [Google Scholar] [CrossRef] [PubMed]
- Katz, J.M.; Webster, R.G. Efficacy of inactivated influenza A virus (H3N2) vaccines grown in mammalian cells or embryonated eggs. J. Infect. Dis. 1989, 160, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Meyer, W.J.; Wood, J.M.; Major, D.; Robertson, J.S.; Webster, R.G.; Katz, J.M. Influence of host cell-mediated variation on the international surveillance of influenza A (H3N2) viruses. Virology 1993, 196, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Ampofo, W.K.; Azziz-Baumgartner, E.; Bashir, U.; Cox, N.J.; Fasce, R.; Giovanni, M.; Grohmann, G.; Huang, S.; Katz, J.; Mironenko, A.; et al. Strengthening the influenza vaccine virus selection and development process. In Presented at the 3rd WHO Informal Consultation for Improving Influenza Vaccine Virus Selection, Geneva, Switzerland, 1–3 April 2014.
- Salk, J.; Salk, D. Control of influenza and poliomyelitis with killed virus vaccines. Science 1977, 195, 834–847. [Google Scholar] [CrossRef] [PubMed]
- De Villiers, P.J.; Steele, A.D.; Hiemstra, L.A.; Rappaport, R.; Dunning, A.J.; Gruber, W.C.; Forrest, B.D. Efficacy and safety of a live-attenuated influenza vaccine in adults 60 years of age and older. Vaccine 2009, 28, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Tlaxca, J.L.; Ellis, S.; Remmele, R.L., Jr. Live attenuated and inactivated viral vaccine formulation and nasal delivery: Potential and challenges. Adv. Drug. Deliv. Rev. 2015, 93, 56–78. [Google Scholar] [CrossRef] [PubMed]
- Laver, W.G.; Webster, R.G. Preparation and immunogenicity of an influenza virus hemagglutinin and neuraminidase subunit vaccine. Virology 1976, 69, 511–522. [Google Scholar] [CrossRef]
- Wong, S.S.; Webby, R.J. Traditional and new influenza vaccines. Clin. Microbiol. Rev. 2013, 26, 476–492. [Google Scholar] [CrossRef] [PubMed]
- Duxbury, A.E.; Hampson, A.W.; Sievers, J.G. Antibody response in humans to deoxycholate-treated influenza virus vaccine. J. Immunol. 1968, 101, 62–67. [Google Scholar] [PubMed]
- Baxter, R.; Patriarca, P.A.; Ensor, K.; Izikson, R.; Goldenthal, K.L.; Cox, M.M. Evaluation of the safety, reactogenicity and immunogenicity of flublok(r) trivalent recombinant baculovirus-expressed hemagglutinin influenza vaccine administered intramuscularly to healthy adults 50–64 years of age. Vaccine 2011, 29, 2272–2278. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. A study of conditions for the optimum production of PR8 influenza virus in chick embryos. J. Exp. Med. 1944, 79, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Maassab, H.F.; Bryant, M.L. The development of live-attenuated cold-adapted influenza virus vaccine for humans. Rev. Med. Virol. 1999, 9, 237–244. [Google Scholar] [CrossRef]
- Snyder, M.H.; Betts, R.F.; DeBorde, D.; Tierney, E.L.; Clements, M.L.; Herrington, D.; Sears, S.D.; Dolin, R.; Maassab, H.F.; Murphy, B.R. Four viral genes independently contribute to attenuation of live influenza A/Ann Arbor/6/60 (H2N2) cold-adapted reassortant virus vaccines. J. Virol. 1988, 62, 488–495. [Google Scholar] [PubMed]
- Cox, N.J.; Kitame, F.; Kendal, A.P.; Maassab, H.F.; Naeve, C. Identification of sequence changes in the cold-adapted, live-attenuated influenza vaccine strain, A/Ann Arbor/6/60 (H2N2). Virology 1988, 167, 554–567. [Google Scholar] [CrossRef]
- Maassab, H.F. Plaque formation of influenza virus at 25 °C. Nature 1968, 219, 645–646. [Google Scholar] [CrossRef] [PubMed]
- Couch, R.B.; Keitel, W.A.; Cate, T.R. Improvement of inactivated influenza virus vaccines. J. Infect. Dis. 1997, 176, S38–S44. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Aspelund, A.; Kemble, G.; Jin, H. Molecular studies of temperature-sensitive replication of the cold-adapted B/Ann Arbor/1/66, the master donor virus for live-attenuated influenza flumist vaccines. Virology 2008, 380, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Mahmood, K.; Chen, Z.; Yang, C.F.; Spaete, J.; Greenberg, H.B.; Herlocher, M.L.; Jin, H.; Kemble, G. Multiple gene segments control the temperature sensitivity and attenuation phenotypes of ca B/Ann Arbor/1/66. J. Virol. 2005, 79, 11014–11021. [Google Scholar] [CrossRef] [PubMed]
- DeBorde, D.C.; Donabedian, A.M.; Herlocher, M.L.; Naeve, C.W.; Maassab, H.F. Sequence comparison of wild-type and cold-adapted B/Ann Arbor/1/66 influenza virus genes. Virology 1988, 163, 429–443. [Google Scholar] [CrossRef]
- Donabedian, A.M.; DeBorde, D.C.; Maassab, H.F. Genetics of cold-adapted B/Ann Arbor/1/66 influenza virus reassortants: The acidic polymerase (PA) protein gene confers temperature sensitivity and attenuated virulence. Microb. Pathog. 1987, 3, 97–108. [Google Scholar] [CrossRef]
- Chan, W.; Zhou, H.; Kemble, G.; Jin, H. The cold adapted and temperature sensitive influenza A/Ann Arbor/6/60 virus, the master donor virus for live-attenuated influenza vaccines, has multiple defects in replication at the restrictive temperature. Virology 2008, 380, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Zhou, H.; Lu, B.; Kemble, G. Imparting temperature sensitivity and attenuation in ferrets to A/Puerto Rico/8/34 influenza virus by transferring the genetic signature for temperature sensitivity from cold-adapted A/Ann Arbor/6/60. J. Virol. 2004, 78, 995–998. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Lu, B.; Zhou, H.; Ma, C.; Zhao, J.; Yang, C.F.; Kemble, G.; Greenberg, H. Multiple amino acid residues confer temperature sensitivity to human influenza virus vaccine strains (flumist) derived from cold-adapted A/Ann Arbor/6/60. Virology 2003, 306, 18–24. [Google Scholar] [CrossRef]
- Maassab, H.F. Adaptation and growth characteristics of influenza virus at 25 °C. Nature 1967, 213, 612–614. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.; Baker, S.F.; Nogales, A.; Martinez-Sobrido, L.; Dewhurst, S. Development of a mouse-adapted live-attenuated influenza virus that permits in vivo analysis of enhancements to the safety of live-attenuated influenza virus vaccine. J. Virol. 2015, 89, 3421–3426. [Google Scholar] [CrossRef] [PubMed]
- Greenhawt, M.J. Influenza vaccination in asthmatic patients. J. Allergy Clin. Immunol. 2014, 133, 1233–1234. [Google Scholar] [CrossRef] [PubMed]
- Belshe, R.B.; Gruber, W.C. Safety, efficacy and effectiveness of cold-adapted, live, attenuated, trivalent, intranasal influenza vaccine in adults and children. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 1947–1951. [Google Scholar] [PubMed]
- Ghendon, Y.Z.; Polezhaev, F.I.; Lisovskaya, K.V.; Medvedeva, T.E.; Alexandrova, G.I.; Klimov, A.I. Recombinant cold-adapted attenuated influenza A vaccines for use in children: Molecular genetic analysis of the cold-adapted donor and recombinants. Infect. Immun. 1984, 44, 730–733. [Google Scholar] [PubMed]
- Ghendon, Y.Z. Molecular characteristics and biological properties (genetic markers) of candidate strains for preparation of live influenza virus vaccines. Bull. World Health Organ. 1984, 62, 493–499. [Google Scholar] [PubMed]
- Isakova-Sivak, I.; Chen, L.M.; Matsuoka, Y.; Voeten, J.T.; Kiseleva, I.; Heldens, J.G.; den Bosch, H.; Klimov, A.; Rudenko, L.; Cox, N.J.; et al. Genetic bases of the temperature-sensitive phenotype of a master donor virus used in live-attenuated influenza vaccines: A/Leningrad/134/17/57 (H2N2). Virology 2011, 412, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Kendal, A.P. Cold-adapted live-attenuated influenza vaccines developed in russia: Can they contribute to meeting the needs for influenza control in other countries? Eur. J. Epidemiol. 1997, 13, 591–609. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.I.; Cordeiro, M.; Sevilla, E.; Liu, J. Comparison of egg and high yielding MDCK cell-derived live-attenuated influenza virus for commercial production of trivalent influenza vaccine: In vitro cell susceptibility and influenza virus replication kinetics in permissive and semi-permissive cells. Vaccine 2010, 28, 3848–3855. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, W.; Zhou, H.; Suguitan, A.L., Jr.; Shambaugh, C.; Kim, L.; Zhao, J.; Kemble, G.; Jin, H. Generation of live-attenuated novel influenza virus A/California/7/09 (H1N1) vaccines with high yield in embryonated chicken eggs. J. Virol. 2010, 84, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Pearson, H.E. Distribution of influenza virus type a in infected eggs and the survival of virus under certain conditions of storage. J. Bacteriol. 1944, 48, 369–378. [Google Scholar] [PubMed]
- Voyles, C. A flu vaccine shortage can be devastating for LTC residents. RN 2005, 68, 10. [Google Scholar] [PubMed]
- Stulc, J.P. The politics of the flu vaccine shortage. J. Ky. Med. Assoc. 2004, 102, 563–564. [Google Scholar] [PubMed]
- Genzel, Y.; Reichl, U. Continuous cell lines as a production system for influenza vaccines. Expert Rev. Vaccines 2009, 8, 1681–1692. [Google Scholar] [CrossRef] [PubMed]
- Le Ru, A.; Jacob, D.; Transfiguracion, J.; Ansorge, S.; Henry, O.; Kamen, A.A. Scalable production of influenza virus in HEK-293 cells for efficient vaccine manufacturing. Vaccine 2010, 28, 3661–3671. [Google Scholar] [CrossRef] [PubMed]
- Merten, O.W.; Manuguerra, J.C.; Hannoun, C.; van der Werf, S. Production of influenza virus in serum-free mammalian cell cultures. Dev. Biol. Stand. 1999, 98, 23–37. [Google Scholar] [PubMed]
- Merten, O.W.; Hannoun, C.; Manuguerra, J.C.; Ventre, F.; Petres, S. Production of influenza virus in cell cultures for vaccine preparation. Adv. Exp. Med. Biol. 1996, 397, 141–151. [Google Scholar] [PubMed]
- Del Giudice, G.; Rappuoli, R. Inactivated and adjuvanted influenza vaccines. Curr. Top. Microbiol. Immunol. 2015, 386, 151–180. [Google Scholar] [PubMed]
- Wu, J.; Liu, S.Z.; Dong, S.S.; Dong, X.P.; Zhang, W.L.; Lu, M.; Li, C.G.; Zhou, J.C.; Fang, H.H.; Liu, Y.; et al. Safety and immunogenicity of adjuvanted inactivated split-virion and whole-virion influenza A (H5N1) vaccines in children: A phase I-II randomized trial. Vaccine 2010, 28, 6221–6227. [Google Scholar] [CrossRef] [PubMed]
- Black, S. Safety and effectiveness of MF-59 adjuvanted influenza vaccines in children and adults. Vaccine 2015, 33, B3–B5. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Jackson, L.A.; Edwards, K.M.; Keitel, W.A.; Hill, H.; Noah, D.L.; Creech, C.B.; Patel, S.M.; Mangal, B.; Kotloff, K.L. Persistence of antibody to influenza A/H5N1 vaccine virus: Impact of AS03 adjuvant. Clin. Vaccine. Immunol. 2016, 23, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Stobart, C.C.; Moore, M.L. RNA virus reverse genetics and vaccine design. Viruses 2014, 6, 2531–2550. [Google Scholar] [CrossRef] [PubMed]
- Almazan, F.; Marquez-Jurado, S.; Nogales, A.; Enjuanes, L. Engineering infectious cDNAs of coronavirus as bacterial artificial chromosomes. Methods Mol. Biol. 2015, 1282, 135–152. [Google Scholar] [PubMed]
- Almazan, F.; Dediego, M.L.; Galan, C.; Escors, D.; Alvarez, E.; Ortego, J.; Sola, I.; Zuniga, S.; Alonso, S.; Moreno, J.L.; et al. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 2006, 80, 10900–10906. [Google Scholar] [CrossRef] [PubMed]
- Racaniello, V.R.; Baltimore, D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science 1981, 214, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Sobrido, L.; Garcia-Sastre, A. Generation of recombinant influenza virus from plasmid DNA. J. Vis. Exp. 2010, 42, e2057. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, O.G. Many ways to make an influenza virus—Review of influenza virus reverse genetics methods. Influenza Other Respir. Viruses 2013, 7, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Nogales, A.; Baker, S.F.; Ortiz-Riano, E.; Dewhurst, S.; Topham, D.J.; Martinez-Sobrido, L. Influenza a virus attenuation by codon deoptimization of the NS gene for vaccine development. J. Virol. 2014, 88, 10525–10540. [Google Scholar] [CrossRef] [PubMed]
- Nogales, A.; Baker, S.F.; Martinez-Sobrido, L. Replication-competent influenza A viruses expressing a red fluorescent protein. Virology 2014, 476C, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Sobrido, L.; Cadagan, R.; Steel, J.; Basler, C.F.; Palese, P.; Moran, T.M.; Garcia-Sastre, A. Hemagglutinin-pseudotyped green fluorescent protein-expressing influenza viruses for the detection of influenza virus neutralizing antibodies. J. Virol. 2010, 84, 2157–2163. [Google Scholar] [CrossRef] [PubMed]
- Nogales, A.; Baker, S.F.; Domm, W.; Martinez-Sobrido, L. Development and applications of single-cycle infectious influenza A virus (sciIAV). Virus Res. 2015, 216, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Breen, M.; Nogales, A.; Baker, S.F.; Martinez-Sobrido, L. Replication-competent influenza A viruses expressing reporter genes. Viruses 2016, 8, 179. [Google Scholar] [CrossRef] [PubMed]
- Nogales, A.; Rodriguez-Sanchez, I.; Monte, K.; Lenschow, D.J.; Perez, D.R.; Martinez-Sobrido, L. Replication-competent fluorescent-expressing influenza B virus. Virus Res. 2015, 213, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Breen, M.; Nogales, A.; Baker, S.F.; Perez, D.R.; Martinez-Sobrido, L. Replication-competent influenza A and B viruses expressing a fluorescent dynamic timer protein for in vitro and in vivo studies. PLoS ONE 2016, 11, e0147723. [Google Scholar] [CrossRef] [PubMed]
- Zobel, A.; Neumann, G.; Hobom, G. RNA polymerase I catalysed transcription of insert viral cDNA. Nucleic Acids Res. 1993, 21, 3607–3614. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Zobel, A.; Hobom, G. RNA polymerase I-mediated expression of influenza viral RNA molecules. Virology 1994, 202, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Neumann, G.; Kawaoka, Y.; Hobom, G.; Webster, R.G. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 2000, 97, 6108–6113. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Neumann, G.; Hobom, G.; Webster, R.G.; Kawaoka, Y. “Ambisense” approach for the generation of influenza A virus: vRNA and mRNA synthesis from one template. Virology 2000, 267, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.Y.; Ortiz-Riano, E.; de la Torre, J.C.; Martinez-Sobrido, L. Generation of recombinant arenavirus for vaccine development in fda-approved vero cells. J. Vis. Exp. 2013, 78, e50662. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Riano, E.; Cheng, B.Y.; Carlos de la Torre, J.; Martinez-Sobrido, L. Arenavirus reverse genetics for vaccine development. J. Gen. Virol. 2013, 94, 1175–1188. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Watanabe, T.; Ito, H.; Watanabe, S.; Goto, H.; Gao, P.; Hughes, M.; Perez, D.R.; Donis, R.; Hoffmann, E.; et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 1999, 96, 9345–9350. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Duke, G.M. Cloning of the canine RNA polymerase I promoter and establishment of reverse genetics for influenza A and B in MDCK cells. Virol. J. 2007, 4, 102. [Google Scholar] [CrossRef] [PubMed]
- Massin, P.; Rodrigues, P.; Marasescu, M.; van der Werf, S.; Naffakh, N. Cloning of the chicken RNA polymerase I promoter and use for reverse genetics of influenza A viruses in avian cells. J. Virol. 2005, 79, 13811–13816. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Curtiss, R., 3rd. Efficient generation of influenza virus with a mouse RNA polymerase I-driven all-in-one plasmid. Virol. J. 2015, 12, 95. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; He, D.; Wang, Z.; Ou, S.; Yuan, R.; Li, S. Cloning the horse RNA polymerase I promoter and its application to studying influenza virus polymerase activity. Viruses 2016, 8, 119. [Google Scholar] [CrossRef] [PubMed]
- Fodor, E.; Devenish, L.; Engelhardt, O.G.; Palese, P.; Brownlee, G.G.; Garcia-Sastre, A. Rescue of influenza A virus from recombinant DNA. J. Virol. 1999, 73, 9679–9682. [Google Scholar] [PubMed]
- Zhou, B.; Lin, X.; Wang, W.; Halpin, R.A.; Bera, J.; Stockwell, T.B.; Barr, I.G.; Wentworth, D.E. Universal influenza B virus genomic amplification facilitates sequencing, diagnostics, and reverse genetics. J. Clin. Microbiol. 2014, 52, 1330–1337. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.; Cadman, A.; Zurcher, T.; Barclay, W.S. A reverse genetics approach for recovery of recombinant influenza B viruses entirely from cDNA. J. Virol. 2002, 76, 11744–11747. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Mahmood, K.; Yang, C.F.; Webster, R.G.; Greenberg, H.B.; Kemble, G. Rescue of influenza B virus from eight plasmids. Proc. Natl. Acad. Sci. USA 2002, 99, 11411–11416. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, M.; Kawaoka, Y. Taming influenza viruses. Virus Res. 2011, 162, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Marsh, G.A.; Hatami, R.; Palese, P. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J. Virol. 2007, 81, 9727–9736. [Google Scholar] [CrossRef] [PubMed]
- Isel, C.; Munier, S.; Naffakh, N. Experimental approaches to study genome packaging of influenza A viruses. Viruses 2016, 8, 218. [Google Scholar] [CrossRef] [PubMed]
- Schickli, J.H.; Flandorfer, A.; Nakaya, T.; Martinez-Sobrido, L.; Garcia-Sastre, A.; Palese, P. Plasmid-only rescue of influenza A virus vaccine candidates. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 1965–1973. [Google Scholar] [PubMed]
- Steel, J.; Lowen, A.C.; Pena, L.; Angel, M.; Solorzano, A.; Albrecht, R.; Perez, D.R.; Garcia-Sastre, A.; Palese, P. Live attenuated influenza viruses containing NS1 truncations as vaccine candidates against H5N1 highly pathogenic avian influenza. J. Virol. 2009, 83, 1742–1753. [Google Scholar] [CrossRef] [PubMed]
- Richt, J.A.; Garcia-Sastre, A. Attenuated influenza virus vaccines with modified NS1 proteins. Curr. Top. Microbiol. Immunol. 2009, 333, 177–195. [Google Scholar] [PubMed]
- Vincent, A.L.; Ma, W.; Lager, K.M.; Janke, B.H.; Webby, R.J.; Garcia-Sastre, A.; Richt, J.A. Efficacy of intranasal administration of a truncated NS1 modified live influenza virus vaccine in swine. Vaccine 2007, 25, 7999–8009. [Google Scholar] [CrossRef] [PubMed]
- Quinlivan, M.; Zamarin, D.; Garcia-Sastre, A.; Cullinane, A.; Chambers, T.; Palese, P. Attenuation of equine influenza viruses through truncations of the NS1 protein. J. Virol. 2005, 79, 8431–8439. [Google Scholar] [CrossRef] [PubMed]
- Falcon, A.M.; Fernandez-Sesma, A.; Nakaya, Y.; Moran, T.M.; Ortin, J.; Garcia-Sastre, A. Attenuation and immunogenicity in mice of temperature-sensitive influenza viruses expressing truncated NS1 proteins. J. Gen. Virol. 2005, 86, 2817–2821. [Google Scholar] [CrossRef] [PubMed]
- Ferko, B.; Stasakova, J.; Romanova, J.; Kittel, C.; Sereinig, S.; Katinger, H.; Egorov, A. Immunogenicity and protection efficacy of replication-deficient influenza A viruses with altered NS1 genes. J. Virol. 2004, 78, 13037–13045. [Google Scholar] [CrossRef] [PubMed]
- Richt, J.A.; Lekcharoensuk, P.; Lager, K.M.; Vincent, A.L.; Loiacono, C.M.; Janke, B.H.; Wu, W.H.; Yoon, K.J.; Webby, R.J.; Solorzano, A.; et al. Vaccination of pigs against swine influenza viruses by using an NS1-truncated modified live-virus vaccine. J. Virol. 2006, 80, 11009–11018. [Google Scholar] [CrossRef] [PubMed]
- Solorzano, A.; Webby, R.J.; Lager, K.M.; Janke, B.H.; Garcia-Sastre, A.; Richt, J.A. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 2005, 79, 7535–7543. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Suarez, D.L.; Pantin-Jackwood, M.; Mibayashi, M.; Garcia-Sastre, A.; Saif, Y.M.; Lee, C.W. Characterization of influenza virus variants with different sizes of the non-structural (NS) genes and their potential as a live influenza vaccine in poultry. Vaccine 2008, 26, 3580–3586. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.H.; Song, M.S.; Park, S.J.; Pascua, P.N.; Baek, Y.H.; Kwon, H.I.; Kim, E.H.; Kim, S.; Jang, H.K.; Poo, H.; et al. Development of a dual-protective live-attenuated vaccine against H5N1 and H9N2 avian influenza viruses by modifying the NS1 gene. Arch. Virol. 2015, 160, 1729–1740. [Google Scholar] [CrossRef] [PubMed]
- Nogales, A.; Huang, K.; Chauche, C.; DeDiego, M.L.; Murcia, P.R.; Parrish, C.R.; Martinez-Sobrido, L. Canine influenza viruses with modified NS1 proteins for the development of live-attenuated vaccines. Virology 2016, 500, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Baskin, C.R.; Bielefeldt-Ohmann, H.; Garcia-Sastre, A.; Tumpey, T.M.; van Hoeven, N.; Carter, V.S.; Thomas, M.J.; Proll, S.; Solorzano, A.; Billharz, R.; et al. Functional genomic and serological analysis of the protective immune response resulting from vaccination of macaques with an NS1-truncated influenza virus. J. Virol. 2007, 81, 11817–11827. [Google Scholar] [CrossRef] [PubMed]
- Pica, N.; Langlois, R.A.; Krammer, F.; Margine, I.; Palese, P. NS1-truncated live-attenuated virus vaccine provides robust protection to aged mice from viral challenge. J. Virol. 2012, 86, 10293–10301. [Google Scholar] [CrossRef] [PubMed]
- Hai, R.; Martinez-Sobrido, L.; Fraser, K.A.; Ayllon, J.; Garcia-Sastre, A.; Palese, P. Influenza B virus NS1-truncated mutants: Live-attenuated vaccine approach. J. Virol. 2008, 82, 10580–10590. [Google Scholar] [CrossRef] [PubMed]
- Talon, J.; Salvatore, M.; O'Neill, R.E.; Nakaya, Y.; Zheng, H.; Muster, T.; Garcia-Sastre, A.; Palese, P. Influenza A and B viruses expressing altered NS1 proteins: A vaccine approach. Proc. Natl. Acad. Sci. USA 2000, 97, 4309–4314. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.Y.; Ortiz-Riano, E.; Nogales, A.; de la Torre, J.C.; Martinez-Sobrido, L. Development of live-attenuated arenavirus vaccines based on codon deoptimization. J. Virol. 2015, 89, 3523–3533. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.; Coleman, J.R.; Papamichail, D.; Ward, C.B.; Nimnual, A.; Futcher, B.; Skiena, S.; Wimmer, E. Live attenuated influenza virus vaccines by computer-aided rational design. Nat. Biotechnol. 2010, 28, 723–726. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Skiena, S.; Futcher, B.; Mueller, S.; Wimmer, E. Deliberate reduction of hemagglutinin and neuraminidase expression of influenza virus leads to an ultraprotective live vaccine in mice. Proc. Natl. Acad. Sci. USA 2013, 110, 9481–9486. [Google Scholar] [CrossRef] [PubMed]
- Katsura, H.; Iwatsuki-Horimoto, K.; Fukuyama, S.; Watanabe, S.; Sakabe, S.; Hatta, Y.; Murakami, S.; Shimojima, M.; Horimoto, T.; Kawaoka, Y. A replication-incompetent virus possessing an uncleavable hemagglutinin as an influenza vaccine. Vaccine 2012, 30, 6027–6033. [Google Scholar] [CrossRef] [PubMed]
- Powell, T.J.; Silk, J.D.; Sharps, J.; Fodor, E.; Townsend, A.R. Pseudotyped influenza A virus as a vaccine for the induction of heterotypic immunity. J. Virol. 2012, 86, 13397–13406. [Google Scholar] [CrossRef] [PubMed]
- Shinya, K.; Fujii, Y.; Ito, H.; Ito, T.; Kawaoka, Y. Characterization of a neuraminidase-deficient influenza A virus as a potential gene delivery vector and a live vaccine. J. Virol. 2004, 78, 3083–3088. [Google Scholar] [CrossRef] [PubMed]
- Uraki, R.; Kiso, M.; Iwatsuki-Horimoto, K.; Fukuyama, S.; Takashita, E.; Ozawa, M.; Kawaoka, Y. A novel bivalent vaccine based on a PB2-knockout influenza virus protects mice from pandemic H1N1 and highly pathogenic H5N1 virus challenges. J. Virol. 2013, 87, 7874–7881. [Google Scholar] [CrossRef] [PubMed]
- Victor, S.T.; Watanabe, S.; Katsura, H.; Ozawa, M.; Kawaoka, Y. A replication-incompetent PB2-knockout influenza A virus vaccine vector. J. Virol. 2012, 86, 4123–4128. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Baker, S.F.; Martinez-Sobrido, L.; Topham, D.J. Induction of CD8 T cell heterologous protection by a single dose of single-cycle infectious influenza virus. J. Virol. 2014, 88, 12006–12016. [Google Scholar] [CrossRef] [PubMed]
- Masic, A.; Pyo, H.M.; Babiuk, S.; Zhou, Y. An eight-segment swine influenza virus harboring H1 and H3 hemagglutinins is attenuated and protective against H1N1 and H3N2 subtypes in pigs. J. Virol. 2013, 87, 10114–10125. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, W.; Ozawa, M.; Hatta, M.; Orozco, E.; Martinez, M.B.; Kawaoka, Y. A recombinant influenza virus vaccine expressing the f protein of respiratory syncytial virus. Arch. Virol. 2014, 159, 1067–1077. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Iwatsuki-Horimoto, K.; Kiso, M.; Uraki, R.; Ichiko, Y.; Takimoto, T.; Kawaoka, Y. A replication-incompetent influenza virus bearing the hn glycoprotein of human parainfluenza virus as a bivalent vaccine. Vaccine 2013, 31, 6239–6246. [Google Scholar] [CrossRef] [PubMed]
- Katsura, H.; Piao, Z.; Iwatsuki-Horimoto, K.; Akeda, Y.; Watanabe, S.; Horimoto, T.; Oishi, K.; Kawaoka, Y. A bivalent vaccine based on a replication-incompetent influenza virus protects against streptococcus pneumoniae and influenza virus infection. J. Virol. 2014, 88, 13410–13417. [Google Scholar] [CrossRef] [PubMed]
- Pena, L.; Sutton, T.; Chockalingam, A.; Kumar, S.; Angel, M.; Shao, H.; Chen, H.; Li, W.; Perez, D.R. Influenza viruses with rearranged genomes as live-attenuated vaccines. J. Virol. 2013, 87, 5118–5127. [Google Scholar] [CrossRef] [PubMed]
- Nogales, A.; DeDiego, M.L.; Topham, D.J.; Martinez-Sobrido, L. Rearrangement of influenza virus spliced segments for the development of live-attenuated vaccines. J. Virol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Perez, D.R.; Garcia-Sastre, A. H5N1, a wealth of knowledge to improve pandemic preparedness. Virus Res. 2013, 178, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Perez, D.R.; Lim, W.; Seiler, J.P.; Yi, G.; Peiris, M.; Shortridge, K.F.; Webster, R.G. Role of quail in the interspecies transmission of H9 influenza A viruses: Molecular changes on HA that correspond to adaptation from ducks to chickens. J. Virol. 2003, 77, 3148–3156. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Cho, K.J.; Fiers, W.; Saelens, X. M2e-based universal influenza A vaccines. Vaccines 2015, 3, 105–136. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I.A.; Skehel, J.J.; Wiley, D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 1981, 289, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, L.; Compans, R.W.; Wang, B.Z. Universal influenza vaccines, a dream to be realized soon. Viruses 2014, 6, 1974–1991. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wittman, V.; Byers, A.; Tapia, T.; Zhou, B.; Warren, W.; Heaton, P.; Connolly, K. In vitro stimulation of human influenza-specific CD8+ T cells by dendritic cells pulsed with an influenza virus-like particle (VLP) vaccine. Vaccine 2010, 28, 5524–5532. [Google Scholar] [CrossRef] [PubMed]
- Latham, T.; Galarza, J.M. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J. Virol. 2001, 75, 6154–6165. [Google Scholar] [CrossRef] [PubMed]
- Pushko, P.; Tumpey, T.M.; Bu, F.; Knell, J.; Robinson, R.; Smith, G. Influenza virus-like particles comprised of the HA, NA, and M1 proteins of H9N2 influenza virus induce protective immune responses in BALB/c mice. Vaccine 2005, 23, 5751–5759. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.J.; Leser, G.P.; Morita, E.; Lamb, R.A. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J. Virol. 2007, 81, 7111–7123. [Google Scholar] [CrossRef] [PubMed]
- Pushko, P.; Pearce, M.B.; Ahmad, A.; Tretyakova, I.; Smith, G.; Belser, J.A.; Tumpey, T.M. Influenza virus-like particle can accommodate multiple subtypes of hemagglutinin and protect from multiple influenza types and subtypes. Vaccine 2011, 29, 5911–5918. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nogales, A.; Martínez-Sobrido, L. Reverse Genetics Approaches for the Development of Influenza Vaccines. Int. J. Mol. Sci. 2017, 18, 20. https://doi.org/10.3390/ijms18010020
Nogales A, Martínez-Sobrido L. Reverse Genetics Approaches for the Development of Influenza Vaccines. International Journal of Molecular Sciences. 2017; 18(1):20. https://doi.org/10.3390/ijms18010020
Chicago/Turabian StyleNogales, Aitor, and Luis Martínez-Sobrido. 2017. "Reverse Genetics Approaches for the Development of Influenza Vaccines" International Journal of Molecular Sciences 18, no. 1: 20. https://doi.org/10.3390/ijms18010020
APA StyleNogales, A., & Martínez-Sobrido, L. (2017). Reverse Genetics Approaches for the Development of Influenza Vaccines. International Journal of Molecular Sciences, 18(1), 20. https://doi.org/10.3390/ijms18010020