TGF-β and Physiological Root Resorption of Deciduous Teeth
Abstract
:1. Introduction
2. Results
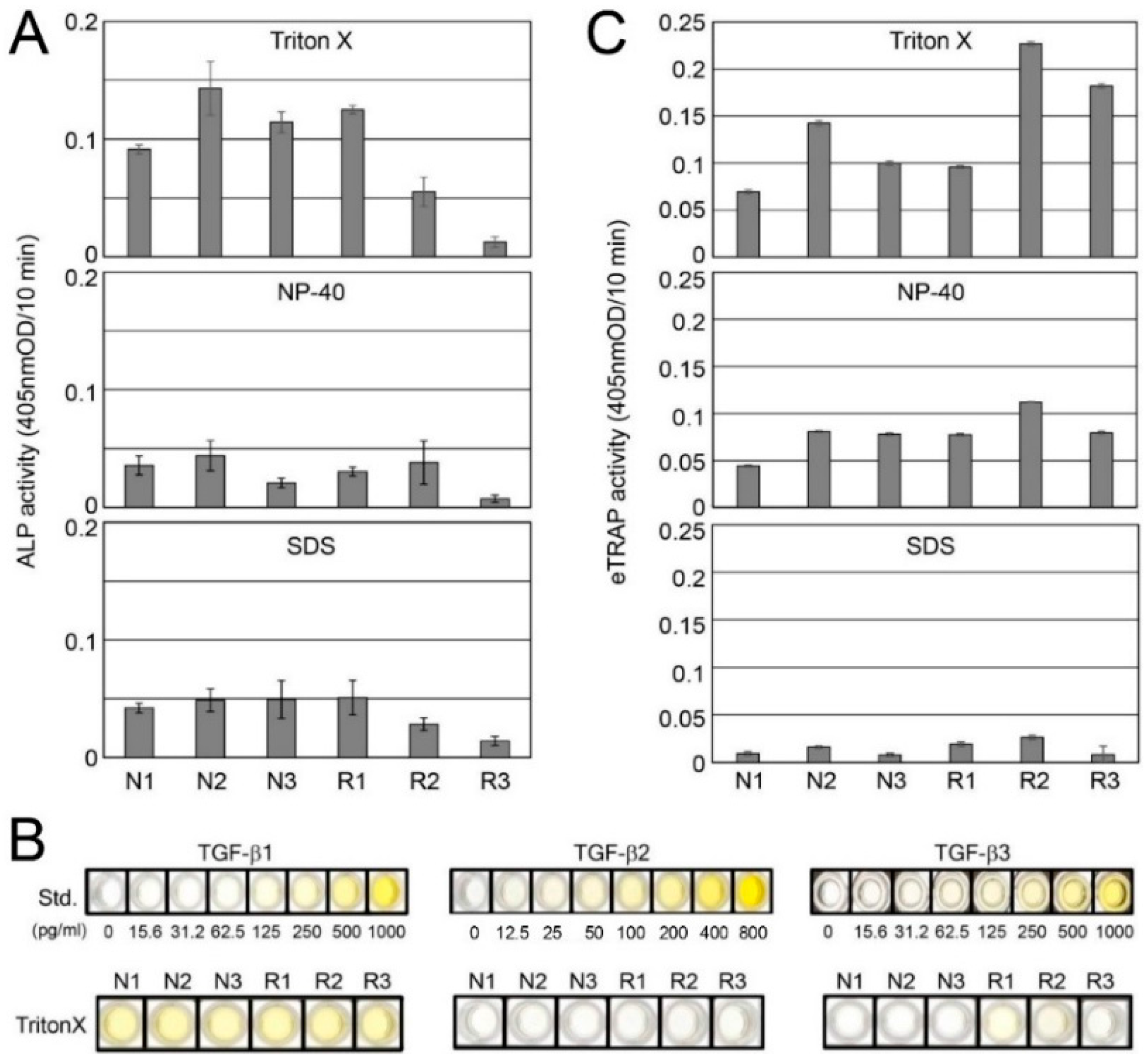
2.1. Sequential Extraction of Proteins from Root-Surrounding Tissues
2.2. Detection of TGF-β and Tartrate-Resistant Acid Phosphatase (TRAP) Activities in Root-Surrounding Tissues and Identification of TGF-β Isoforms
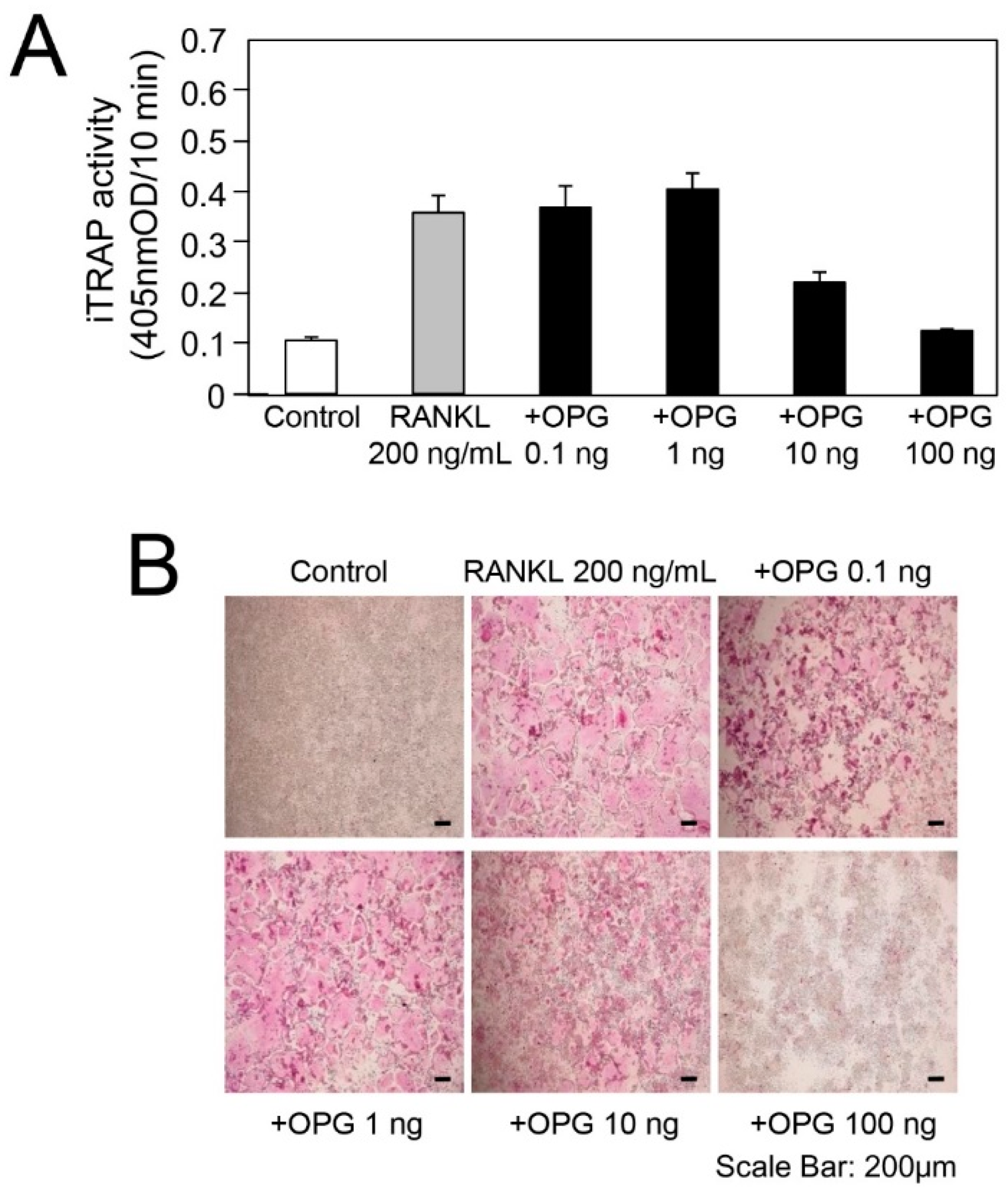
2.3. In Vitro Experiment for RANKL-Mediated Osteoclast Differentiation
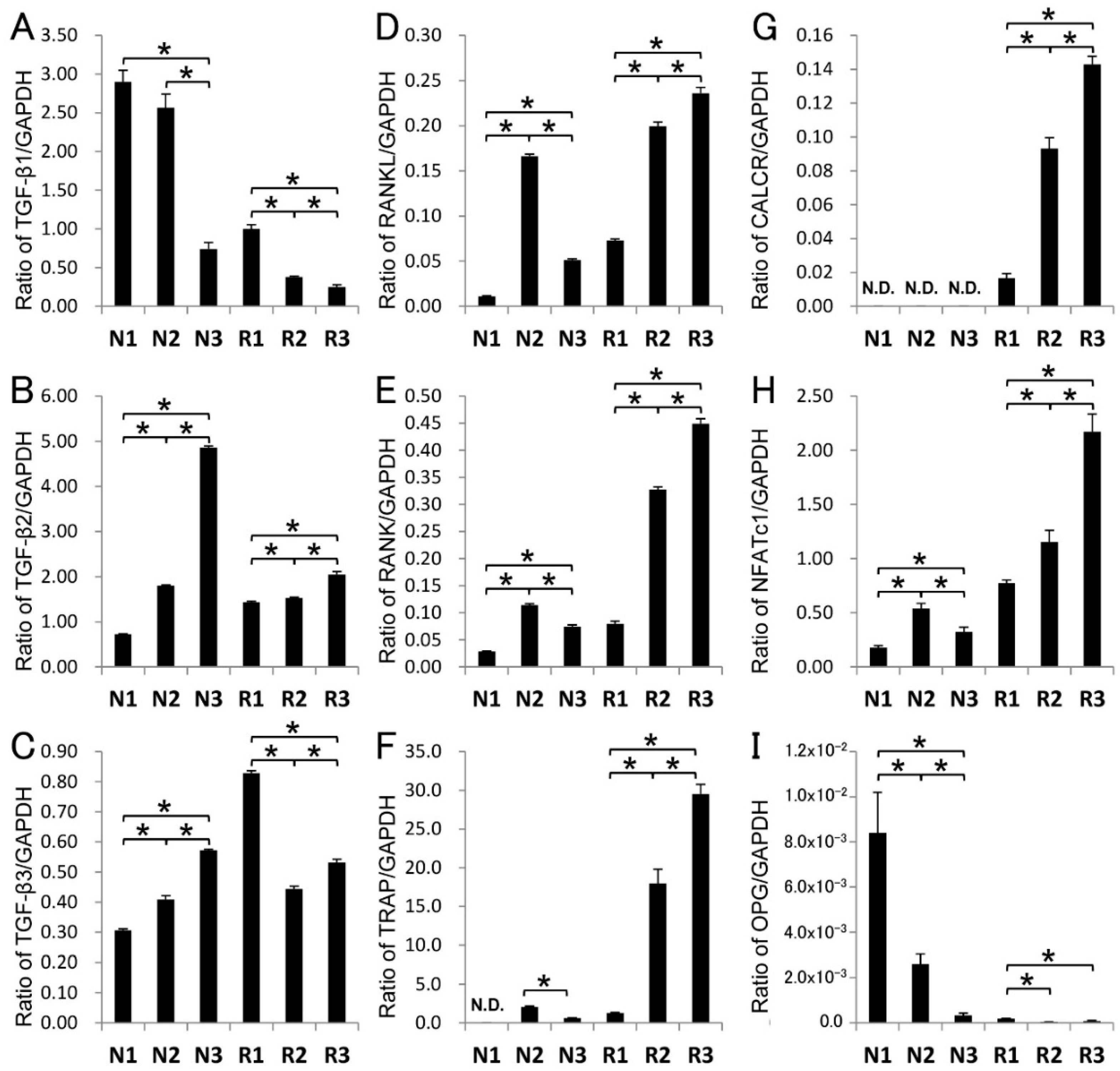
2.4. Gene Expression in Root-Surrounding Tissues
3. Discussion
3.1. TGF-β in Root-Surrounding Tissues of Deciduous Teeth
3.2. TGF-β Activity in Root-Surrounding Tissues of Deciduous Teeth and In Vitro Experiment for RANKL-Mediated Osteoclast Differentiation
3.3. Potential Roles of TGF-β in Root-Surrounding Tissues of Deciduous Teeth
4. Materials and Methods
4.1. Tissue Preparation
4.2. Sequential Extraction of Proteins from Root-Surrounding Tissues
4.3. Alkaline Phosphatase (ALP) Activity Assay (ALP-HPDL System)
4.4. SDS-PAGE
4.5. Endogenous and Induced Tartrate-Resistant Acid Phosphatase (TRAP) Assays and TRAP Staining
4.6. Enzyme-Linked Immunosorbent Assay (ELISA)
4.7. Quantitative Real-Time PCR (qPCR)
4.8. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
| Triton X | Tissue (mg) | 660 Assay (μg/mL) | Total (μg/μL) | μg/mg Tissue |
|---|---|---|---|---|
| N1 | 106.3 | 5450 | 2779.5/510 | 26.1 |
| N2 | 99.45 | 4700 | 3543.8/754 | 35.6 |
| N3 | 109.2 | 6500 | 4680/720 | 42.9 |
| R1 | 109.36 | 3875 | 2828.8/730 | 25.9 |
| R2 | 102.8 | 6125 | 3711.8/606 | 36.1 |
| R3 | 156.7 | 5450 | 2550.6/468 | 16.3 |
| NP40 | Tissue (mg) | 660 Assay (μg/mL) | Total (μg/µL) | μg/mg Tissue |
| N1 | 106.3 | 1210 | 750.2/620 | 7.06 |
| N2 | 99.45 | 1330 | 771.4/580 | 7.76 |
| N3 | 109.2 | 1630 | 717.2/440 | 6.57 |
| R1 | 109.36 | 1200 | 648/540 | 5.93 |
| R2 | 102.8 | 1190 | 747.3/628 | 7.27 |
| R3 | 156.7 | 960 | 604.8/630 | 3.86 |
| SDS | Tissue (mg) | 660 Assay (μg/mL) | Total (μg/μL) | μg/mg Tissue |
| N1 | 106.3 | 135 | 82.4/610 | 0.78 |
| N2 | 99.45 | 120 | 70.8/590 | 0.71 |
| N3 | 109.2 | 100 | 56.2/562 | 0.51 |
| R1 | 109.36 | 135 | 79.7/590 | 0.73 |
| R2 | 102.8 | 145 | 67.9/468 | 0.66 |
| R3 | 156.7 | 85 | 39.3/462 | 0.25 |
| Gene | Sequence (5′–3′) | Size (bp) | qPCR Protocol (45 Cycles) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Denaturation | Annealing | Extension | |||||||
| °C | Second | °C | Second | °C | Second | ||||
| TGF-β1 | Forward | TTCATGAACCCAAGGGCTACC | 104 | 95 | 10 | 65 | 10 | 72 | 15 |
| Reverse | CTGGTTGTACAGAGCCAGGAC | ||||||||
| TGF-β2 | Forward | CAGTGGGAAGACCCCACATC | 150 | 60 | |||||
| Reverse | ATGTAAAGTGGACGCAGGCA | ||||||||
| TGF-β3 | Forward | CTGTGCGTGAATGGCTCTTG | 93 | ||||||
| Reverse | TATCCCCGTTGGGCTGAAAG | ||||||||
| RANKL | Forward | ACGATCAACGCCACAGACAT | 89 | 65 | |||||
| Reverse | TTGGAGATCTTGGCCCAACC | ||||||||
| RANK | Forward | GAAGCACGTCATGGGACTGA | 95 | 60 | |||||
| Reverse | CTGGGCAAGTAAGTCTGGGG | ||||||||
| TRAP | Forward | AACGTCTCGGCACAGATAGC | 101 | 65 | |||||
| Reverse | GACACATTGGACCGTGGGAT | ||||||||
| CALCR | Forward | CGCCTGTGGTGGTATCATGT | 142 | ||||||
| Reverse | CAGGGCCGTGGATGATGTAA | ||||||||
| NFATc1 | Forward | TCGTGGAGAAAGCACCAGAC | 91 | ||||||
| Reverse | TCGACCACCAAGGAATTCGG | ||||||||
| OPG | Forward | TGTTCTGGAAACAGTGAATCGAC | 80 | ||||||
| Reverse | ACAGCAAACCTGAAGAACGC | ||||||||
| GAPDH | Forward | CCATCACCATCTTCCAGGAG | 346 | ||||||
| Reverse | ACAGTCTTCTGGGTGGCAGT | ||||||||
Abbreviations
| SDS | Sodium dodecyl sulfate |
| PAGE | Polyacrylamide gel electrophoresis |
| NP-40 | Nonidet P-40 |
| kDa | Killodalton |
| ACP5 | Acid phosphatase 5 |
| PCR | Polymerase chain reaction |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| GST | Glutathione S-transferase |
References
- Ten Cate, A.R.; Anderson, R.D. An ultrastructural study of tooth resorption in the kitten. J. Dent. Res. 1986, 65, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Motegi, N.; Suzuki, H.; Watanabe, C.; Tadokoro, K.; Yanagisawa, T.; Higashi, S. Dentin resorption mediated by odontoclasts in physiological root resorption of human deciduous teeth. Am. J. Anat. 1988, 183, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Takahashi, N.; Higashi, S.; Suda, T. Multinucleated cells formed on calcified dentine from mouse bone marrow cells treated with 1α,25-dihydroxyvitamin D3 have ruffled borders and resorb dentine. Anat. Rec. 1989, 224, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Addison, W.C. Enzyme histochemical characteristics of human and kitten odontoclasts and kitten osteoclasts: A comparative study using whole cells. Histochem. J. 1979, 11, 719–735. [Google Scholar] [CrossRef] [PubMed]
- Oshiro, T.; Shibasaki, Y.; Martin, T.J.; Sasaki, T. Immunolocalization of vacuolar-type H+-ATPase, cathepsin K, matrix metalloproteinase-9, and receptor activator of NF-κB ligand in odontoclasts during physiological root resorption of human deciduous teeth. Anat. Rec. 2001, 264, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, H.; Shima, N.; Nakagawa, N.; Yamaguchi, K.; Kinosaki, M.; Mochizuki, S.; Tomoyasu, A.; Yano, K.; Goto, M.; Murakami, A.; et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Nat. Acad. Sci. USA 1998, 95, 3597–3602. [Google Scholar] [CrossRef] [PubMed]
- Simonet, W.S.; Lacey, D.L.; Dunstan, C.R.; Kelley, M.; Chang, M.S.; Luthy, R.; Nguyen, H.Q.; Wooden, S.; Bennett, L.; Boone, T.; et al. Osteoprotegerin: A novel secreted protein involved in the regulation of bone density. Cell 1997, 89, 309–319. [Google Scholar] [CrossRef]
- Kanzaki, H.; Chiba, M.; Shimizu, Y.; Mitani, H. Dual regulation of osteoclast differentiation by periodontal ligament cells through RANKL stimulation and OPG inhibition. J. Dent. Res. 2001, 80, 887–891. [Google Scholar] [CrossRef] [PubMed]
- Boabaid, F.; Berry, J.E.; Koh, A.J.; Somerman, M.J.; McCcauley, L.K. The role of parathyroid hormone-related protein in the regulation of osteoclastogenesis by cementoblasts. J. Periodontol. 2004, 75, 1247–1254. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, R.; Usui, M.; Yamamoto, G.; Nishii, K.; Tsukamoto, Y.; Okamatsu, Y.; Sato, T.; Asou, Y.; Nakashima, K.; Yamamoto, M. Tumor necrosis factor-α enhances RANKL expression in gingival epithelial cells via protein kinase a signaling. J. Periodontal Res. 2014, 49, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Thirunavukkarasu, K.; Miles, R.R.; Halladay, D.L.; Yang, X.; Galvin, R.J.; Chandrasekhar, S.; Martin, T.J.; Onyia, J.E. Stimulation of osteoprotegerin (OPG) gene expression by transforming growth factor-β (TGF-β). Mapping of the opg promoter region that mediates TGF-β effects. J. Biol. Chem. 2001, 276, 36241–36250. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Shang, L.; Chen, X.; Kong, X.; Liu, N.; Bai, Y.; Fang, J.; Dang, J.; Wang, X.; Jin, Y. Deciduous dental pulp stem cells are involved in osteoclastogenesis during physiologic root resorption. J. Cell. Physiol. 2013, 228, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Wise, G.E.; Frazier-Bowers, S.; D′Souza, R.N. Cellular, molecular, and genetic determinants of tooth eruption. Crit. Rev. Oral Biol. Med. 2002, 13, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Nakchbandi, I.A.; Weir, E.E.; Insogna, K.L.; Philbrick, W.M.; Broadus, A.E. Parathyroid hormone-related protein induces spontaneous osteoclast formation via a paracrine cascade. Proc. Nat. Acad. Sci. USA 2000, 97, 7296–7300. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, Y. Growth and differentiation factors exist in porcine periodontal ligament tissues. J. Jpn. Soc. Periodontol. 2010, 52, 24–36. [Google Scholar] [CrossRef]
- Huang, T.; Schor, S.L.; Hinck, A.P. Biological activity differences between TGF-β1 and TGF-β3 correlate with differences in the rigidity and arrangement of their component monomers. Biochemistry 2014, 53, 5737–5749. [Google Scholar] [CrossRef] [PubMed]
- Takai, H.; Kanematsu, M.; Yano, K.; Tsuda, E.; Higashio, K.; Ikeda, K.; Watanabe, K.; Yamada, Y. Transforming growth factor-β stimulates the production of osteoprotegerin/osteoclastogenesis inhibitory factor by bone marrow stromal cells. J. Biol. Chem. 1998, 273, 27091–27096. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Yamamoto, M.; Ono, K.; Nishikawa, M.; Nagata, N.; Motoyoshi, K.; Akatsu, T. Transforming growth factor-β1 increases mrna levels of osteoclastogenesis inhibitory factor in osteoblastic/stromal cells and inhibits the survival of murine osteoclast-like cells. Biochem. Biophys. Res. Commun. 1998, 252, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Sells Galvin, R.J.; Gatlin, C.L.; Horn, J.W.; Fuson, T.R. TGF-β enhances osteoclast differentiation in hematopoietic cell cultures stimulated with RANKL and M-CSF. Biochem. Biophys. Res. Commun. 1999, 265, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.M.; Itoh, K.; Udagawa, N.; Hausler, K.; Yasuda, H.; Shima, N.; Mizuno, A.; Higashio, K.; Takahashi, N.; Suda, T.; et al. Transforming growth factor β affects osteoclast differentiation via direct and indirect actions. J. Bone Miner. Res. 2001, 16, 1787–1794. [Google Scholar] [CrossRef] [PubMed]
- Koseki, T.; Gao, Y.; Okahashi, N.; Murase, Y.; Tsujisawa, T.; Sato, T.; Yamato, K.; Nishihara, T. Role of TGF-β family in osteoclastogenesis induced by RANKL. Cell Signal. 2002, 14, 31–36. [Google Scholar] [CrossRef]
- Yasui, T.; Kadono, Y.; Nakamura, M.; Oshima, Y.; Matsumoto, T.; Masuda, H.; Hirose, J.; Omata, Y.; Yasuda, H.; Imamura, T.; et al. Regulation of RANKL-induced osteoclastogenesis by TGF-β through molecular interaction between SMAD3 and TRAF6. J. Bone Miner. Res. 2011, 26, 1447–1456. [Google Scholar] [CrossRef] [PubMed]
- Ota, K.; Quint, P.; Ruan, M.; Pederson, L.; Westendorf, J.J.; Khosla, S.; Oursler, M.J. TGF-β induces Wnt10b in osteoclasts from female mice to enhance coupling to osteoblasts. Endocrinology 2013, 154, 3745–3752. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi-Kinoshita, S.; Yamakoshi, Y.; Onuma, K.; Yamamoto, R.; Asada, Y. TGF-β1 autocrine signalling and enamel matrix components. Sci. Rep. 2016, 6, 33644. [Google Scholar] [CrossRef] [PubMed]
- Yamakoshi, Y.; Kinoshita, S.; Izuhara, L.; Karakida, T.; Fukae, M.; Oida, S. DPP and DSP are necessary for maintaining TGF-β1 activity in dentin. J. Dent. Res. 2014, 93, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, A.; Romaris, M.; Rasmussen, L.M.; Heinegard, D.; Twardzik, D.R.; Border, W.A.; Ruoslahti, E. Interaction of the small interstitial proteoglycans biglycan, decorin and fibromodulin with transforming growth factor β. Biochem. J. 1994, 302, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Iozzo, R.V.; Murdoch, A.D. Proteoglycans of the extracellular environment: Clues from the gene and protein side offer novel perspectives in molecular diversity and function. FASEB J. 1996, 10, 598–614. [Google Scholar] [PubMed]
- Hyytiainen, M.; Penttinen, C.; Keski-Oja, J. Latent TGF-β binding proteins: Extracellular matrix association and roles in TGF-β activation. Crit. Rev. Clin. Lab. Sci. 2004, 41, 233–264. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.M.; Sugars, R.V.; Wendel, M.; Smith, A.J.; Waddington, R.J.; Cooper, P.R.; Sloan, A.J. TGF-β/extracellular matrix interactions in dentin matrix: A role in regulating sequestration and protection of bioactivity. Calcif. Tissue Int. 2009, 85, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Schonherr, E.; Broszat, M.; Brandan, E.; Bruckner, P.; Kresse, H. Decorin core protein fragment Leu155-Val260 interacts with TGF-β but does not compete for decorin binding to type I collagen. Arch. Biochem. Biophys. 1998, 355, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Massague, J. Receptors for the TGF-β family. Cell 1992, 69, 1067–1070. [Google Scholar] [CrossRef]
- Kingsley, D.M. The TGF-β superfamily: New members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 1994, 8, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, Y.; Hiraide, S.; Iizuka, K. Isoform-specific regulation of transforming growth factor-β mRNA expression in macrophages in response to adrenoceptor stimulation. Microbiol. Immunol. 2016, 60, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Inman, G.J.; Nicolas, F.J.; Callahan, J.F.; Harling, J.D.; Gaster, L.M.; Reith, A.D.; Laping, N.J.; Hill, C.S. Sb-431542 is a potent and specific inhibitor of transforming growth factor-β superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002, 62, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Laping, N.J.; Grygielko, E.; Mathur, A.; Butter, S.; Bomberger, J.; Tweed, C.; Martin, W.; Fornwald, J.; Lehr, R.; Harling, J.; et al. Inhibition of transforming growth factor (TGF)-β1-induced extracellular matrix with a novel inhibitor of the TGF-β type I receptor kinase activity: Sb-431542. Mol. Pharmacol. 2002, 62, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lu, Y.; Dai, J.; Yao, Z.; Kitazawa, R.; Kitazawa, S.; Zhao, X.; Hall, D.E.; Pienta, K.J.; Keller, E.T. In vivo real-time imaging of TGF-β-induced transcriptional activation of the RANK ligand gene promoter in intraosseous prostate cancer. Prostate 2004, 59, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Ishida, A.; Fujita, N.; Kitazawa, R.; Tsuruo, T. Transforming growth factor-β induces expression of receptor activator of NF-κB ligand in vascular endothelial cells derived from bone. J. Biol. Chem. 2002, 277, 26217–26224. [Google Scholar] [CrossRef] [PubMed]
- Eeles, D.G.; Hodge, J.M.; Singh, P.P.; Schuijers, J.A.; Grills, B.L.; Gillespie, M.T.; Myers, D.E.; Quinn, J.M. Osteoclast formation elicited by interleukin-33 stimulation is dependent upon the type of osteoclast progenitor. Mol. Cell. Endocrinol. 2015, 399, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.F.; Zhang, Y.K.; Yu, Z.S.; Zhou, J.L. The role of the serum RANKL/OPG ratio in the healing of intertrochanteric fractures in elderly patients. Mol. Med. Rep. 2013, 7, 1169–1172. [Google Scholar] [PubMed]
- Nagano, T.; Oida, S.; Suzuki, S.; Iwata, T.; Yamakoshi, Y.; Ogata, Y.; Gomi, K.; Arai, T.; Fukae, M. Porcine enamel protein fractions contain transforming growth factor-β1. J. Periodontol. 2006, 77, 1688–1694. [Google Scholar] [CrossRef] [PubMed]
- Ujiie, Y.; Karakida, T.; Yamakoshi, Y.; Ohshima, T.; Gomi, K.; Oida, S. Interleukin-4 released from human gingival fibroblasts reduces osteoclastogenesis. Arch. Oral Biol. 2016, 72, 187–193. [Google Scholar] [CrossRef] [PubMed]


| Root-Surrounding Tissue | Non-Resorption | Resorption | |||||
|---|---|---|---|---|---|---|---|
| Cervical | Center | Apical | Cervical | Center | Apical | ||
| Region | N1 | N2 | N3 | R1 | R2 | R3 | |
| Protein study | Collagen | ↑ | ↑↑ | ↑↑↑ | ↑↑↑ | ↑↑ | trace |
| Decorin | ↑ | ↑↑↑ | ↑↑ | ↑↑ | ↑ | trace | |
| ALP activity | ↑ | ↑↑ | ↑↑ | ↑↑ | ↑ | trace | |
| eTRAP activity | ↑ | ↑ | ↑↑ | ↑↑ | ↑↑↑↑ | ↑↑↑ | |
| In vitro study | iTRAP activity | ↑ | ↑↑ | ↑ | ↑ | trace | trace |
| Genetic study (mRNA level) | TGF-β1 | ↑↑↑↑ | ↑↑↑↑ | ↑↑ | ↑↑ | ↑ | ↑ |
| TGF-β2 | ↑ | ↑↑ | ↑↑↑↑ | ↑ | ↑ | ↑↑ | |
| TGF-β3 | ↑ | ↑↑ | ↑↑↑ | ↑↑↑ | ↑ | ↑↑ | |
| RANKL | trace | ↑↑↑ | ↑ | ↑ | ↑↑↑ | ↑↑↑↑ | |
| RANK | trace | ↑↑ | ↑ | ↑ | ↑↑↑ | ↑↑↑↑ | |
| TRAP | N.D | ↑ | trace | ↑ | ↑↑↑ | ↑↑↑↑ | |
| CALCR | N.D | N.D | N.D | ↑ | ↑↑↑ | ↑↑↑↑ | |
| NFATc1 | trace | ↑↑ | ↑ | ↑↑ | ↑↑↑ | ↑↑↑↑ | |
| OPG | ↑↑↑↑ | ↑↑ | trace | trace | trace | trace | |
| RANKL/OPG ratio | 1.34 | 63.8 | 157 | 393 | 5192 | 3092 | |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shimazaki, E.; Karakida, T.; Yamamoto, R.; Kobayashi, S.; Fukae, M.; Yamakoshi, Y.; Asada, Y. TGF-β and Physiological Root Resorption of Deciduous Teeth. Int. J. Mol. Sci. 2017, 18, 49. https://doi.org/10.3390/ijms18010049
Shimazaki E, Karakida T, Yamamoto R, Kobayashi S, Fukae M, Yamakoshi Y, Asada Y. TGF-β and Physiological Root Resorption of Deciduous Teeth. International Journal of Molecular Sciences. 2017; 18(1):49. https://doi.org/10.3390/ijms18010049
Chicago/Turabian StyleShimazaki, Emi, Takeo Karakida, Ryuji Yamamoto, Saeko Kobayashi, Makoto Fukae, Yasuo Yamakoshi, and Yoshinobu Asada. 2017. "TGF-β and Physiological Root Resorption of Deciduous Teeth" International Journal of Molecular Sciences 18, no. 1: 49. https://doi.org/10.3390/ijms18010049
APA StyleShimazaki, E., Karakida, T., Yamamoto, R., Kobayashi, S., Fukae, M., Yamakoshi, Y., & Asada, Y. (2017). TGF-β and Physiological Root Resorption of Deciduous Teeth. International Journal of Molecular Sciences, 18(1), 49. https://doi.org/10.3390/ijms18010049






