Transcriptional Regulation and Transport of Terpenoid Indole Alkaloid in Catharanthus roseus: Exploration of New Research Directions
Abstract
:1. Introduction
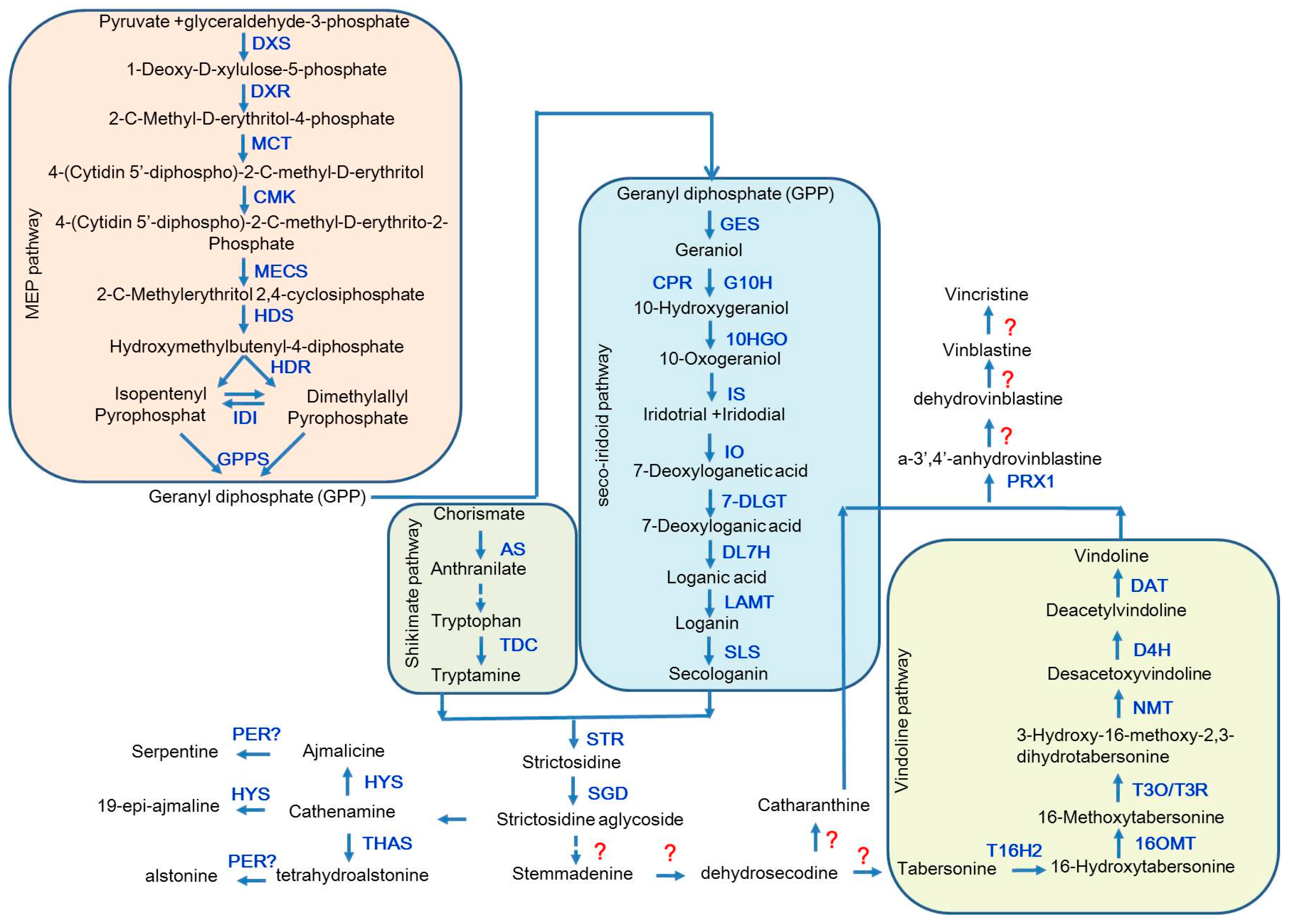
2. The Biosynthetic Pathway of the TIAs in Catharanthus roseus
3. Transcriptional Regulation of TIA Biosynthesis
4. Transport of TIAs in and between Organs, Tissues, Cells and Subcellular Compartments
5. Inter-Cellular Transport Is Required for Biosynthesis of TIAs
6. Intra-Cellular Transport of TIA Intermediates and End Products
7. The Biochemical, Molecular Biological Aspects of TIA Transporters
8. Transcriptomic Data Mining for Exploration of Putative Transcription Factors Involved in TIA Biosynthesis
9. Perspectives and Future Directions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zhao, J.; Dixon, R.A. Mate transporters facilitate vacuolar uptake of epicatechin 3′-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and arabidopsis. Plant Cell 2009, 21, 2323–2340. [Google Scholar] [CrossRef] [PubMed]
- Yazaki, K. Abc transporters involved in the transport of plant secondary metabolites. FEBS Lett. 2006, 580, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J. Flavonoid transport mechanisms: How to go, and with whom. Trends Plant Sci. 2015, 20, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Van Der Heijden, R.; Jacobs, D.; Snoeijer, W.; Hallard, D.; Verpoorte, R. The catharanthus alkaloids: Pharmacognosy and biotechnology. Curr. Med. Chem. 2004, 11, 607. [Google Scholar] [CrossRef] [PubMed]
- Leveque, D.; Jehl, F. Molecular pharmacokinetics of catharanthus (vinca) alkaloids. J. Clin. Pharmacol. 2007, 47, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhang, W.; Zhang, Y.; Chen, J.; Chen, Z. Identification and quantification of active alkaloids in Catharanthus roseus by liquid chromatography-ion trap mass spectrometry. Food Chem. 2013, 139, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.H.; Jin, H.B.; Chen, Y.H.; Cui, L.J.; Ren, W.W.; Gong, Y.F.; Tang, K.X. Terpenoid indole alkaloids biosynthesis and metabolic engineering in Catharanthus roseus. J. Integr. Plant Biol. 2007, 49, 961–974. [Google Scholar] [CrossRef]
- Memelink, J.; Gantet, P. Transcription factors involved in terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Phytochem. Rev. 2007, 6, 353–362. [Google Scholar] [CrossRef]
- Peebles, C.A.; Sander, G.W.; Hughes, E.H.; Peacock, R.; Shanks, J.V.; San, K.Y. The expression of 1-deoxy-d-xylulose synthase and geraniol-10-hydroxylase or anthranilate synthase increases terpenoid indole alkaloid accumulation in Catharanthus roseus hairy roots. Metab. Eng. 2011, 13, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Q.; Fang, X.; Wu, X.M.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Transcriptional regulation of plant secondary metabolism. J. Integr. Plant Biol. 2012, 54, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Courdavault, V.; Papon, N.; Clastre, M.; Giglioli-Guivarc’h, N.; St-Pierre, B.; Burlat, V. A look inside an alkaloid multisite plant: The catharanthus logistics. Curr. Opin. Plant Biol. 2014, 19, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Kellner, F.; Kim, J.; Clavijo, B.J.; Hamilton, J.P.; Childs, K.L.; Vaillancourt, B.; Cepela, J.; Habermann, M.; Steuernagel, B.; Clissold, L. Genome-guided investigation of plant natural product biosynthesis. Plant J. 2015, 82, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Hughes, E.H.; Hong, S.B.; Gibson, S.I.; Shanks, J.V.; San, K.Y. Metabolic engineering of the indole pathway in Catharanthus roseus hairy roots and increased accumulation of tryptamine and serpentine. Metab. Eng. 2004, 6, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.; Qiu, F.; Chen, M.; Zeng, L.; Liu, X.; Yang, C.; Lan, X.; Wang, Q.; Liao, Z. Engineering the mep pathway enhanced ajmalicine biosynthesis. Biotechnol. Appl. Biochem. 2014, 61, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Krithika, R.; Srivastava, P.L.; Rani, B.; Kolet, S.P.; Chopade, M.; Soniya, M.; Thulasiram, H.V. Characterization of 10-hydroxygeraniol dehydrogenase from Catharanthus roseus reveals cascaded enzymatic activity in iridoid biosynthesis. Sci. Rep. 2015, 5, 8258. [Google Scholar] [CrossRef] [PubMed]
- Geu-Flores, F.; Sherden, N.H.; Courdavault, V.; Burlat, V.; Glenn, W.S.; Wu, C.; Nims, E.; Cui, Y.; O’Connor, S.E. An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature 2012, 492, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, K.; Dong, L.; Navrot, N.; Schneider, T.; Burlat, V.; Pollier, J.; Woittiez, L.; van der Krol, S.; Lugan, R.; Ilc, T. The seco-iridoid pathway from Catharanthus roseus. Nat. Commun. 2014, 5, 3606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geerlings, A.; Ibañez, M.M.-L.; Memelink, J.; van der Heijden, R.; Verpoorte, R. Molecular cloning and analysis of strictosidine β-d-glucosidase, an enzyme in terpenoid indole alkaloid biosynthesis in Catharanthus roseus. J. Biol. Chem. 2000, 275, 3051–3056. [Google Scholar] [CrossRef] [PubMed]
- Stavrinides, A.; Tatsis, E.C.; Caputi, L.; Foureau, E.; Stevenson, C.E.; Lawson, D.M.; Courdavault, V.; O’Connor, S.E. Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity. Nat. Commun. 2016, 7, 12116. [Google Scholar] [CrossRef] [PubMed]
- Blom, T.; Sierra, M.; van Vliet, T.; Franke-van Dijk, M.; de Koning, P.; van Iren, F.; Verpoorte, R.; Libbenga, K. Uptake and accumulation of ajmalicine into isolated vacuoles of cultured cells of Catharanthus roseus (L.) G. Don. And its conversion into serpentine. Planta 1991, 183, 170–177. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, M.; Choi, Y.H.; Frederich, M.; Roytrakul, S.; Verpoorte, R. Alkaloid accumulation in Catharanthus roseus cell suspension cultures fed with stemmadenine. Biotechnol. Lett. 2004, 26, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.M.; Hilliou, F.; Duarte, P.; Pereira, L.G.; Almeida, I.; Leech, M.; Memelink, J.; Barcelo, A.R.; Sottomayor, M. Molecular cloning and characterization of a vacuolar class III peroxidase involved in the metabolism of anticancer alkaloids in Catharanthus roseus. Plant Physiol. 2008, 146, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Verpoorte, R. Manipulating indole alkaloid production by Catharanthus roseus cell cultures in bioreactors: From biochemical processing to metabolic engineering. Phytochem. Rev. 2007, 6, 435–457. [Google Scholar] [CrossRef]
- Goossens, A.; Mertens, J.; Pollier, J.; Bossche, R.V.; López-Vidriero, I.; Franco-Zorrilla, J.M. The bhlh transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in Medicago truncatula. Plant Physiol. 2016, 170, 194–210. [Google Scholar]
- Mishra, S.; Phukan, U.J.; Tripathi, V.; Singh, D.K.; Luqman, S.; Shukla, R.K. PsAP2 an AP2/ERF family transcription factor from Papaver somniferum enhances abiotic and biotic stress tolerance in transgenic tobacco. Plant Mol. Biol. 2015, 89, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Li, X.; Kim, S.J.; Kim, H.H.; Lee, J.; Kim, H.; Park, S.U. Myb transcription factors regulate glucosinolate biosynthesis in different organs of chinese cabbage (Brassica rapa ssp. Pekinensis). Molecules 2013, 18, 8682–8695. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.X.; Li, J.X.; Yang, C.Q.; Hu, W.L.; Wang, L.J.; Chen, X.Y. The jasmonate-responsive Ap2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant. 2012, 5, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Menke, F.L.; Champion, A.; Kijne, J.W.; Memelink, J. A novel jasmonate-and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene STR interacts with a jasmonate-and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J. 1999, 18, 4455–4463. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Leopold, A.L.; Sander, G.W.; Shanks, J.V.; Zhao, L.; Gibson, S.I. The ORCA2 transcription factor plays a key role in regulation of the terpenoid indole alkaloid pathway. BMC Plant Biol. 2013, 13, 155. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.-H.; Ren, W.-W.; Cui, L.-J.; Zhang, L.-D.; Sun, X.-F.; Tang, K.-X. Enhanced accumulation of catharanthine and vindoline in Catharanthus roseus hairy roots by overexpression of transcriptional factor ORCA2. Afr. J. Biotechnol. 2011, 10, 3260. [Google Scholar]
- Van der Fits, L.; Memelink, J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 2000, 289, 295–297. [Google Scholar] [CrossRef] [PubMed]
- Peebles, C.A.; Hughes, E.H.; Shanks, J.V.; San, K.Y. Transcriptional response of the terpenoid indole alkaloid pathway to the overexpression of ORCA3 along with jasmonic acid elicitation of Catharanthus roseus hairy roots over time. Metab. Eng. 2009, 11, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Peebles, C.A. Engineering overexpression of ORCA3 and strictosidine glucosidase in Catharanthus roseus hairy roots increases alkaloid production. Protoplasma 2016, 253, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Wang, Q.; Yuan, F.; Xing, S.; Zhao, J.; Choi, Y.H.; Verpoorte, R.; Tian, Y.; Wang, G.; Tang, K. Overexpression of ORCA3 and G10H in Catharanthus roseus plants regulated alkaloid biosynthesis and metabolism revealed by NMR-metabolomics. PLoS ONE 2012, 7, e43038. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zi, J.; Zhu, J.; Chen, S.; Wang, P.; Song, L.; Yu, R. Artemisinic acid serves as a novel ORCA3 inducer to enhance biosynthesis of terpenoid indole alkaloids in Catharanthus roseus cambial meristematic cells. Nat. Prod. Commun. 2016, 11, 715–717. [Google Scholar] [PubMed]
- Pandey, S.S.; Singh, S.; Babu, C.S.; Shanker, K.; Srivastava, N.K.; Shukla, A.K.; Kalra, A. Fungal endophytes of Catharanthus roseus enhance vindoline content by modulating structural and regulatory genes related to terpenoid indole alkaloid biosynthesis. Sci. Rep. 2016, 6, 26583. [Google Scholar] [CrossRef] [PubMed]
- Pré, M.; Sibéril, Y.; Memelink, J.; Champion, A.; Doireau, P.; Gantet, P. Isolation by the yeast one-hybrid system of cdnas encoding transcription factors that bind to the g-box element of the strictosidine synthase gene promoter from Catharanthus roseus. Int. J. Biochromatogr. 2000, 5, 229–244. [Google Scholar]
- Montiel, G.; Zarei, A.; Korbes, A.P.; Memelink, J. The jasmonate-responsive element from the ORCA3 promoter from Catharanthus roseus is active in arabidopsis and is controlled by the transcription factor AtMYC2. Plant Cell Physiol. 2011, 52, 578–587. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hedhili, S.; Montiel, G.; Zhang, Y.; Chatel, G.; Pré, M.; Gantet, P.; Memelink, J. The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 2011, 67, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Van Moerkercke, A.; Steensma, P.; Schweizer, F.; Pollier, J.; Gariboldi, I.; Payne, R.; Bossche, R.V.; Miettinen, K.; Espoz, J.; Purnama, P.C. The bhlh transcription factor bis1 controls the iridoid branch of the monoterpenoid indole alkaloid pathway in Catharanthus roseus. Proc. Natl. Acad. Sci. USA 2015, 112, 8130–8135. [Google Scholar] [CrossRef] [PubMed]
- Van Moerkercke, A.; Steensma, P.; Gariboldi, I.; Espoz, J.; Purnama, P.C.; Schweizer, F.; Miettinen, K.; Vanden Bossche, R.; De Clercq, R.; Memelink, J. The basic helix-loop-helix transcription factor BIS2 is essential for monoterpenoid indole alkaloid production in the medicinal plant Catharanthus roseus. Plant J. 2016, 8, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; Singh, S.K.; Patra, B.; Sui, X.; Pattanaik, S.; Yuan, L. A differentially regulated AP2/ERF transcription factor gene cluster acts downstream of a map kinase cascade to modulate terpenoid indole alkaloid biosynthesis in Catharanthus roseus. New Phytol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Suttipanta, N.; Pattanaik, S.; Kulshrestha, M.; Patra, B.; Singh, S.K.; Yuan, L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011, 157, 2081–2093. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Leopold, A.L.; Sander, G.W.; Shanks, J.V.; Zhao, L.; Gibson, S.I. CrBPF1 overexpression alters transcript levels of terpenoid indole alkaloid biosynthetic and regulatory genes. Front. Plant Sci. 2015, 6, 818. [Google Scholar] [CrossRef] [PubMed]
- Sibéril, Y.; Benhamron, S.; Memelink, J.; Giglioli-Guivarc’h, N.; Thiersault, M.; Boisson, B.; Doireau, P.; Gantet, P. Catharanthus roseus g-box binding factors 1 and 2 act as repressors of strictosidine synthase gene expression in cell cultures. Plant Mol. Biol. 2001, 45, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Pauw, B.; Hilliou, F.A.; Martin, V.S.; Chatel, G.; de Wolf, C.J.; Champion, A.; Pré, M.; van Duijn, B.; Kijne, J.W.; van der Fits, L. Zinc finger proteins act as transcriptional repressors of alkaloid biosynthesis genes in Catharanthus roseus. J. Biol. Chem. 2004, 279, 52940–52948. [Google Scholar] [CrossRef] [PubMed]
- Chebbi, M.; Ginis, O.; Courdavault, V.; Glevarec, G.; Lanoue, A.; Clastre, M.; Papon, N.; Gaillard, C.; Atanassova, R.; St-Pierre, B.; et al. ZCT1 and ZCT2 transcription factors repress the activity of a gene promoter from the methyl erythritol phosphate pathway in madagascar periwinkle cells. J. Plant Physiol. 2014, 171, 1510–1513. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, N.F.; Weaver, J.D.; Cram, E.J.; Lee-Parsons, C.W. Silencing the transcriptional repressor, ZCT1, illustrates the tight regulation of terpenoid indole alkaloid biosynthesis in catharanthus roseus hairy roots. PLoS ONE 2016, 11, e0159712. [Google Scholar] [CrossRef] [PubMed]
- Shitan, N.; Kato, K.; Shoji, T. Alkaloid transporters in plants. Plant Biotechnol. 2014, 31, 453–463. [Google Scholar] [CrossRef]
- Shitan, N.; Yazaki, K. Accumulation and membrane transport of plant alkaloids. Curr. Pharm. Biotechnol. 2007, 8, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Shitan, N.; Bazin, I.; Dan, K.; Obata, K.; Kigawa, K.; Ueda, K.; Sato, F.; Forestier, C.; Yazaki, K. Involvement of cjmdr1, a plant multidrug-resistance-type atp-binding cassette protein, in alkaloid transport in Coptis japonica. Proc. Natl. Acad. Sci. USA 2003, 100, 751–756. [Google Scholar] [CrossRef] [PubMed]
- Shoji, T.; Hashimoto, T. Smoking out the masters: Transcriptional regulators for nicotine biosynthesis in tobacco. Plant Biotechnol. 2013, 30, 217–224. [Google Scholar] [CrossRef]
- Burlat, V.; Oudin, A.; Courtois, M.; Rideau, M.; St-Pierre, B. Co-expression of three mep pathway genes and geraniol 10-hydroxylase in internal phloem parenchyma of Catharanthus roseus implicates multicellular translocation of intermediates during the biosynthesis of monoterpene indole alkaloids and isoprenoid-derived primary metabolites. Plant J. 2004, 38, 131–141. [Google Scholar] [PubMed]
- Verma, P.; Mathur, A.K.; Srivastava, A.; Mathur, A. Emerging trends in research on spatial and temporal organization of terpenoid indole alkaloid pathway in Catharanthus roseus: A literature update. Protoplasma 2012, 249, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Mahroug, S.; Burlat, V.; St-Pierre, B. Cellular and sub-cellular organisation of the monoterpenoid indole alkaloid pathway in Catharanthus roseus. Phytochem. Rev. 2007, 6, 363–381. [Google Scholar] [CrossRef]
- Gutierrez-Nava Mde, L.; Gillmor, C.S.; Jimenez, L.F.; Guevara-Garcia, A.; Leon, P. Chloroplast biogenesis genes act cell and noncell autonomously in early chloroplast development. Plant Physiol. 2004, 135, 471–482. [Google Scholar] [CrossRef] [PubMed]
- De Luca, V.; Salim, V.; Thamm, A.; Masada, S.A.; Yu, F. Making iridoids/secoiridoids and monoterpenoid indole alkaloids: Progress on pathway elucidation. Curr. Opin. Plant Biol. 2014, 19, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Salim, V.; Wiens, B.; Masada-Atsumi, S.; Yu, F.; De Luca, V. 7-deoxyloganetic acid synthase catalyzes a key 3 step oxidation to form 7-deoxyloganetic acid in Catharanthus roseus iridoid biosynthesis. Phytochemistry 2014, 101, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Salim, V.; Yu, F.; Altarejos, J.; Luca, V. Virus-induced gene silencing identifies Catharanthus roseus 7-deoxyloganic acid-7-hydroxylase, a step in iridoid and monoterpene indole alkaloid biosynthesis. Plant J. 2013, 76, 754–765. [Google Scholar] [CrossRef] [PubMed]
- Irmler, S.; Schröder, G.; St-Pierre, B.; Crouch, N.P.; Hotze, M.; Schmidt, J.; Strack, D.; Matern, U.; Schröder, J. Indole alkaloid biosynthesis in Catharanthus roseus: New enzyme activities and identification of cytochrome p450 cyp72a1 as secologanin synthase. Plant J. 2000, 24, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Murata, J.; Roepke, J.; Gordon, H.; de Luca, V. The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell 2008, 20, 524–542. [Google Scholar] [CrossRef] [PubMed]
- Asada, K.; Salim, V.; Masada-Atsumi, S.; Edmunds, E.; Nagatoshi, M.; Terasaka, K.; Mizukami, H.; De Luca, V. A 7-deoxyloganetic acid glucosyltransferase contributes a key step in secologanin biosynthesis in madagascar periwinkle. Plant Cell 2013, 25, 4123–4134. [Google Scholar] [CrossRef] [PubMed]
- Guirimand, G.; Courdavault, V.; Lanoue, A.; Mahroug, S.; Guihur, A.; Blanc, N.; Giglioli-Guivarc’h, N.; St-Pierre, B.; Burlat, V. Strictosidine activation in apocynaceae: Towards a “nuclear time bomb”? BMC Plant Biol. 2010, 10, 182. [Google Scholar] [PubMed]
- Yu, F.; De Luca, V. Atp-binding cassette transporter controls leaf surface secretion of anticancer drug components in Catharanthus roseus. Proc. Natl. Acad. Sci. USA 2013, 110, 15830–15835. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, B.; Vazquez-Flota, F.A.; De Luca, V. Multicellular compartmentation of Catharanthus roseus alkaloid biosynthesis predicts intercellular translocation of a pathway intermediate. Plant Cell 1999, 11, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Easson, M.L.; Froese, J.; Simionescu, R.; Hudlicky, T.; De Luca, V. Completion of the seven-step pathway from tabersonine to the anticancer drug precursor vindoline and its assembly in yeast. Proc. Natl. Acad. Sci. USA 2015, 112, 6224–6229. [Google Scholar] [CrossRef] [PubMed]
- Oudin, A.; Mahroug, S.; Courdavault, V.; Hervouet, N.; Zelwer, C.; Rodríguez-Concepción, M.; St-Pierre, B.; Burlat, V. Spatial distribution and hormonal regulation of gene products from methyl erythritol phosphate and monoterpene-secoiridoid pathways in Catharanthus roseus. Plant Mol. Biol. 2007, 65, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Guirimand, G.; Guihur, A.; Phillips, M.A.; Oudin, A.; Glevarec, G.; Melin, C.; Papon, N.; Clastre, M.; St-Pierre, B.; Rodriguez-Concepcion, M.; et al. A single gene encodes isopentenyl diphosphate isomerase isoforms targeted to plastids, mitochondria and peroxisomes in Catharanthus roseus. Plant Mol. Biol. 2012, 79, 443–459. [Google Scholar] [CrossRef] [PubMed]
- Simkin, A.J.; Miettinen, K.; Claudel, P.; Burlat, V.; Guirimand, G.; Courdavault, V.; Papon, N.; Meyer, S.; Godet, S.; St-Pierre, B. Characterization of the plastidial geraniol synthase from madagascar periwinkle which initiates the monoterpenoid branch of the alkaloid pathway in internal phloem associated parenchyma. Phytochemistry 2013, 85, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Guirimand, G.; Burlat, V.; Oudin, A.; Lanoue, A.; St-Pierre, B.; Courdavault, V. Optimization of the transient transformation of Catharanthus roseus cells by particle bombardment and its application to the subcellular localization of hydroxymethylbutenyl 4-diphosphate synthase and geraniol 10-hydroxylase. Plant Cell Rep. 2009, 28, 1215–1234. [Google Scholar] [CrossRef] [PubMed]
- Guirimand, G.; Guihur, A.; Ginis, O.; Poutrain, P.; Hericourt, F.; Oudin, A.; Lanoue, A.; St-Pierre, B.; Burlat, V.; Courdavault, V. The subcellular organization of strictosidine biosynthesis in Catharanthus roseus epidermis highlights several trans-tonoplast translocations of intermediate metabolites. FEBS J. 2011, 278, 749–763. [Google Scholar] [CrossRef] [PubMed]
- Murata, J.; Luca, V.D. Localization of tabersonine 16-hydroxylase and 16-oh tabersonine-16-O-methyltransferase to leaf epidermal cells defines them as a major site of precursor biosynthesis in the vindoline pathway in Catharanthus roseus. Plant J. 2005, 44, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Besseau, S.; Kellner, F.; Lanoue, A.; Thamm, A.M.; Salim, V.; Schneider, B.; Geu-Flores, F.; Höfer, R.; Guirimand, G.; Guihur, A. A pair of tabersonine 16-hydroxylases initiates the synthesis of vindoline in an organ-dependent manner in Catharanthus roseus. Plant Physiol. 2013, 163, 1792–1803. [Google Scholar] [CrossRef] [PubMed]
- Guirimand, G.; Guihur, A.; Poutrain, P.; Héricourt, F.; Mahroug, S.; St-Pierre, B.; Burlat, V.; Courdavault, V. Spatial organization of the vindoline biosynthetic pathway in Catharanthus roseus. J. Plant Physiol. 2011, 168, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Dethier, M.; De Luca, V. Partial purification of an n-methyltransferase involved in vindoline biosynthesis in Catharanthus roseus. Phytochemistry 1993, 32, 673–678. [Google Scholar] [CrossRef]
- Roytrakul, S.; Verpoorte, R. Role of vacuolar transporter proteins in plant secondary metabolism: Catharanthus roseus cell culture. Phytochem. Rev. 2007, 6, 383–396. [Google Scholar] [CrossRef]
- Roepke, J.; Salim, V.; Wu, M.; Thamm, A.M.; Murata, J.; Ploss, K.; Boland, W.; de Luca, V. Vinca drug components accumulate exclusively in leaf exudates of madagascar periwinkle. Proc. Natl. Acad. Sci. USA 2010, 107, 15287–15292. [Google Scholar] [CrossRef] [PubMed]
- Carqueijeiro, I.; Noronha, H.; Duarte, P.; Gerós, H.; Sottomayor, M. Vacuolar transport of the medicinal alkaloids from Catharanthus roseus is mediated by a proton-driven antiport. Plant Physiol. 2013, 162, 1486–1496. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Hu, Q.; Guo, Y.Q.; Zhu, W.H. Elicitor-induced indole alkaloid biosynthesis in Catharanthus roseus cell cultures is related to Ca2+ influx and the oxidative burst. Plant Sci. 2001, 161, 423–431. [Google Scholar] [CrossRef]
- Lee-Parsons, C.W.; Ertürk, S. Ajmalicine production in methyl jasmonate-induced catharanthus roseus cell cultures depends on Ca2+ level. Plant Cell Rep. 2005, 24, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Davis, L.C.; Verpoorte, R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 2005, 23, 283–333. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Dixon, R.A. The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci. 2010, 15, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Wink, M.; Witte, L. Quinolizidine alkaloids in Genista acanthoclada and its holoparasite, Cuscuta palaestina. J. Chem. Ecol. 1993, 19, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Shitan, N.; Dalmas, F.; Dan, K.; Kato, N.; Ueda, K.; Sato, F.; Forestier, C.; Yazaki, K. Characterization of Coptis japonica CjABCB2, an at p-binding cassette protein involved in alkaloid transport. Phytochemistry 2013, 91, 109–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hildreth, S.B.; Gehman, E.A.; Yang, H.; Lu, R.H.; Ritesh, K.; Harich, K.C.; Yu, S.; Lin, J.; Sandoe, J.L.; Okumoto, S. Tobacco nicotine uptake permease (NUP1) affects alkaloid metabolism. Proc. Natl. Acad. Sci. USA 2011, 108, 18179–18184. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Shitan, N.; Sawada, K.; van Montagu, M.C.; Inzé, D.; Rischer, H.; Goossens, A.; Oksman-Caldentey, K.M.; Moriyama, Y.; Yazaki, K. Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (mate) transporter in Nicotiana tabacum. Proc. Natl. Acad. Sci. USA 2009, 106, 2447–2452. [Google Scholar] [CrossRef] [PubMed]
- Shoji, T.; Inai, K.; Yazaki, Y.; Sato, Y.; Takase, H.; Shitan, N.; Yazaki, K.; Goto, Y.; Toyooka, K.; Matsuoka, K. Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiol. 2009, 149, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Stukkens, Y.; Bultreys, A.; Grec, S.; Trombik, T.; Vanham, D.; Boutry, M. Nppdr1, a pleiotropic drug resistance-type atp-binding cassette transporter from Nicotiana plumbaginifolia, plays a major role in plant pathogen defense. Plant Physiol. 2005, 139, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Pomahačová, B.; Dušek, J.; Dušková, J.; Yazaki, K.; Roytrakul, S.; Verpoorte, R. Improved accumulation of ajmalicine and tetrahydroalstonine in Catharanthus cells expressing an abc transporter. J. Plant Physiol. 2009, 166, 1405–1412. [Google Scholar] [CrossRef] [PubMed]
- Champagne, A.; Rischer, H.; Oksman-Caldentey, K.M.; Boutry, M. In-depth proteome mining of cultured Catharanthus roseus cells identifies candidate proteins involved in the synthesis and transport of secondary metabolites. Proteomics 2012, 12, 3536–3547. [Google Scholar] [CrossRef] [PubMed]
- Deus-Neumann, B.; Zenk, M. A highly selective alkaloid uptake system in vacuoles of higher plants. Planta 1984, 162, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Neumann, D.; Krauss, G.; Hieke, M.; Gröger, D. Indole alkaloid formation and storage in cell suspension cultures of Catharanthus roseus. Planta Med. 1983, 48, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Bessire, M.; Borel, S.; Fabre, G.; Carraça, L.; Efremova, N.; Yephremov, A.; Cao, Y.; Jetter, R.; Jacquat, A.C.; Métraux, J.P. A member of the pleiotropic drug resistance family of atp binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis. Plant Cell 2011, 23, 1958–1970. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Komatsuda, T.; Ma, J.F.; Nawrath, C.; Pourkheirandish, M.; Tagiri, A.; Hu, Y.G.; Sameri, M.; Li, X.; Zhao, X. An atp-binding cassette subfamily g full transporter is essential for the retention of leaf water in both wild barley and rice. Proc. Natl. Acad. Sci. USA 2011, 108, 12354–12359. [Google Scholar] [CrossRef] [PubMed]
- De Geyter, N.; Gholami, A.; Goormachtig, S.; Goossens, A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 2012, 17, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Facchini, P.J.; Bohlmann, J.; Covello, P.S.; de Luca, V.; Mahadevan, R.; Page, J.E.; Ro, D.K.; Sensen, C.W.; Storms, R.; Martin, V.J. Synthetic biosystems for the production of high-value plant metabolites. Trends Biotechnol. 2012, 30, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Zhang, Y.; Chen, X.; Lee, E.J.; Barber, C.J.; Chakrabarty, R.; Desgagné-Penix, I.; Haslam, T.M.; Kim, Y.B.; Liu, E. Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J. Biotechnol. 2013, 166, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.; Ghangal, R.; Sharma, R.; Sinha, A.K.; Jain, M. Transcriptome analysis of Catharanthus roseus for gene discovery and expression profiling. PLoS ONE 2014, 9, e103583. [Google Scholar] [CrossRef] [PubMed]








© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Cai, J.; Wang, R.; Yang, S. Transcriptional Regulation and Transport of Terpenoid Indole Alkaloid in Catharanthus roseus: Exploration of New Research Directions. Int. J. Mol. Sci. 2017, 18, 53. https://doi.org/10.3390/ijms18010053
Liu J, Cai J, Wang R, Yang S. Transcriptional Regulation and Transport of Terpenoid Indole Alkaloid in Catharanthus roseus: Exploration of New Research Directions. International Journal of Molecular Sciences. 2017; 18(1):53. https://doi.org/10.3390/ijms18010053
Chicago/Turabian StyleLiu, Jiaqi, Junjun Cai, Rui Wang, and Shihai Yang. 2017. "Transcriptional Regulation and Transport of Terpenoid Indole Alkaloid in Catharanthus roseus: Exploration of New Research Directions" International Journal of Molecular Sciences 18, no. 1: 53. https://doi.org/10.3390/ijms18010053





