Role of the Genes of Type VI Secretion System in Virulence of Rice Bacterial Brown Stripe Pathogen Acidovorax avenae subsp. avenae Strain RS-2
Abstract
:1. Introduction
2. Results
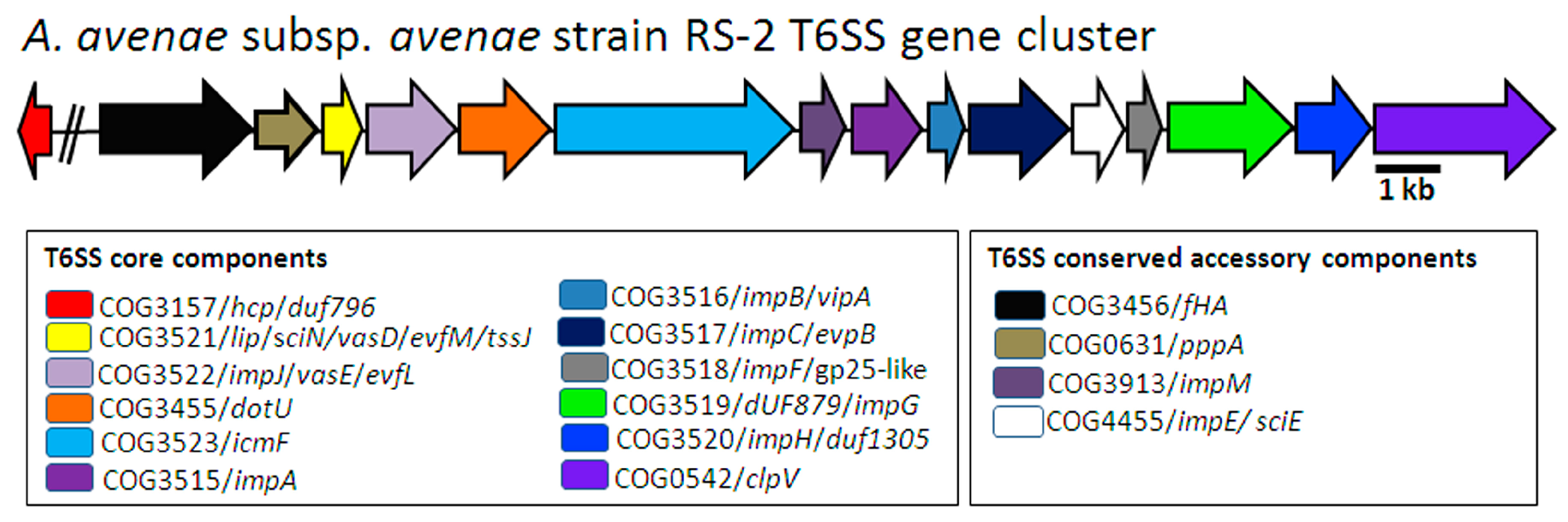
2.1. In Silico Identification of T6SS Genes
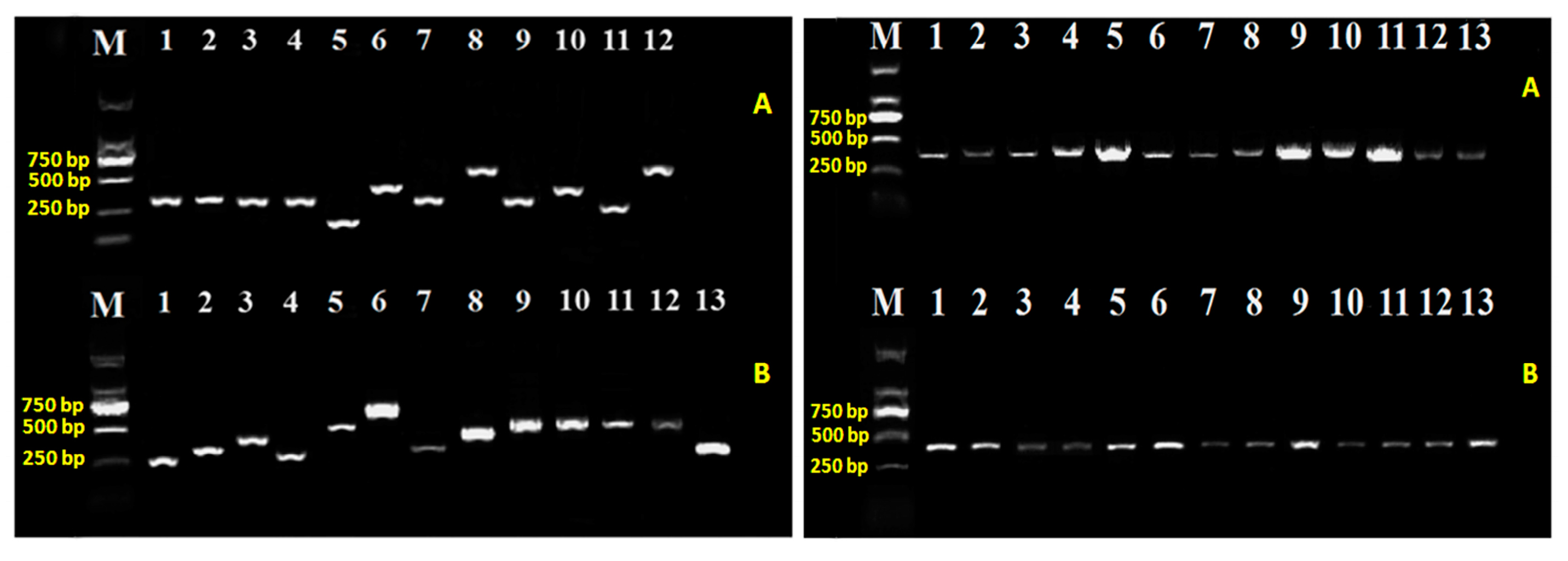
2.2. T6SS Gene Mutants and Complementation
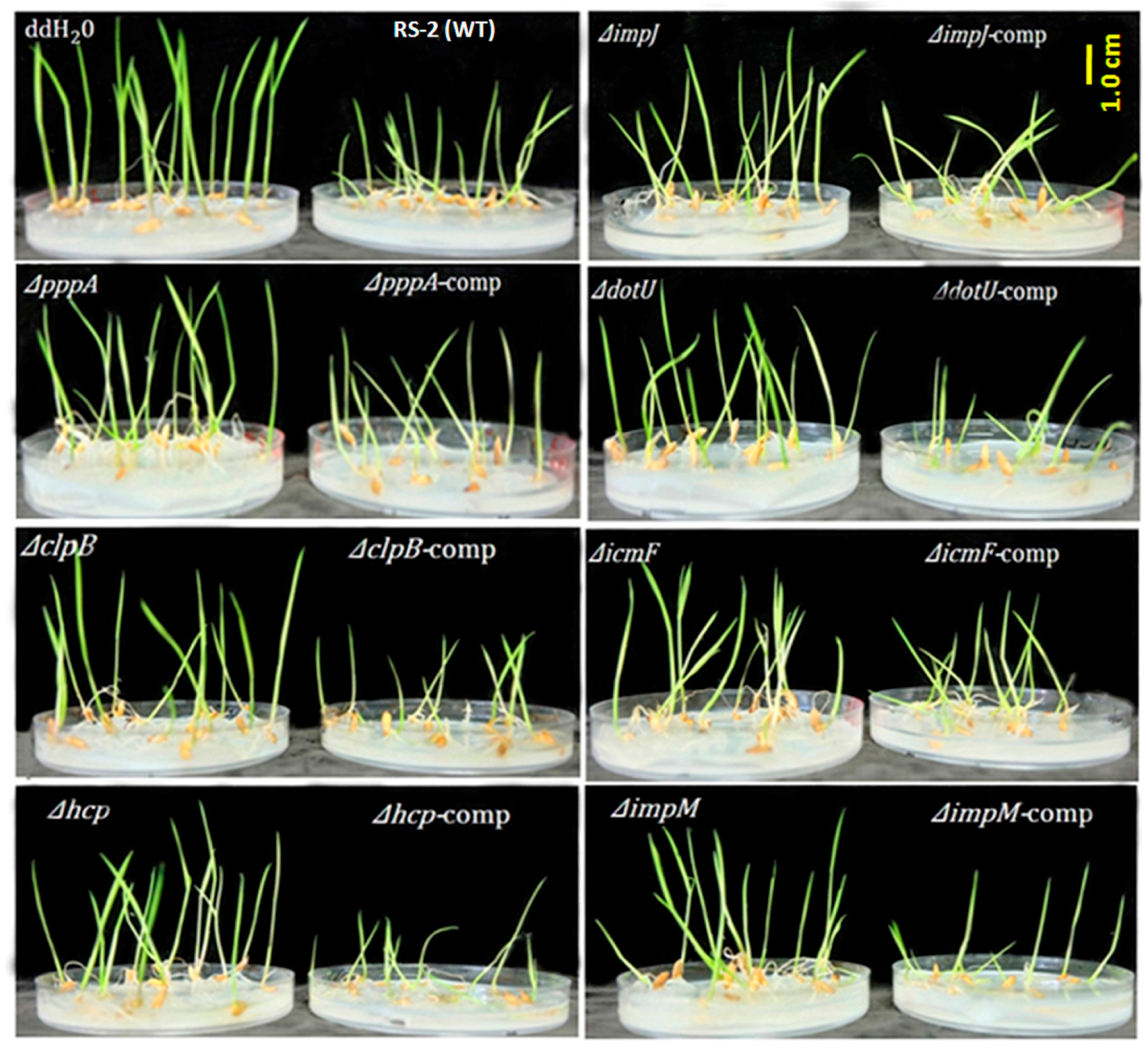
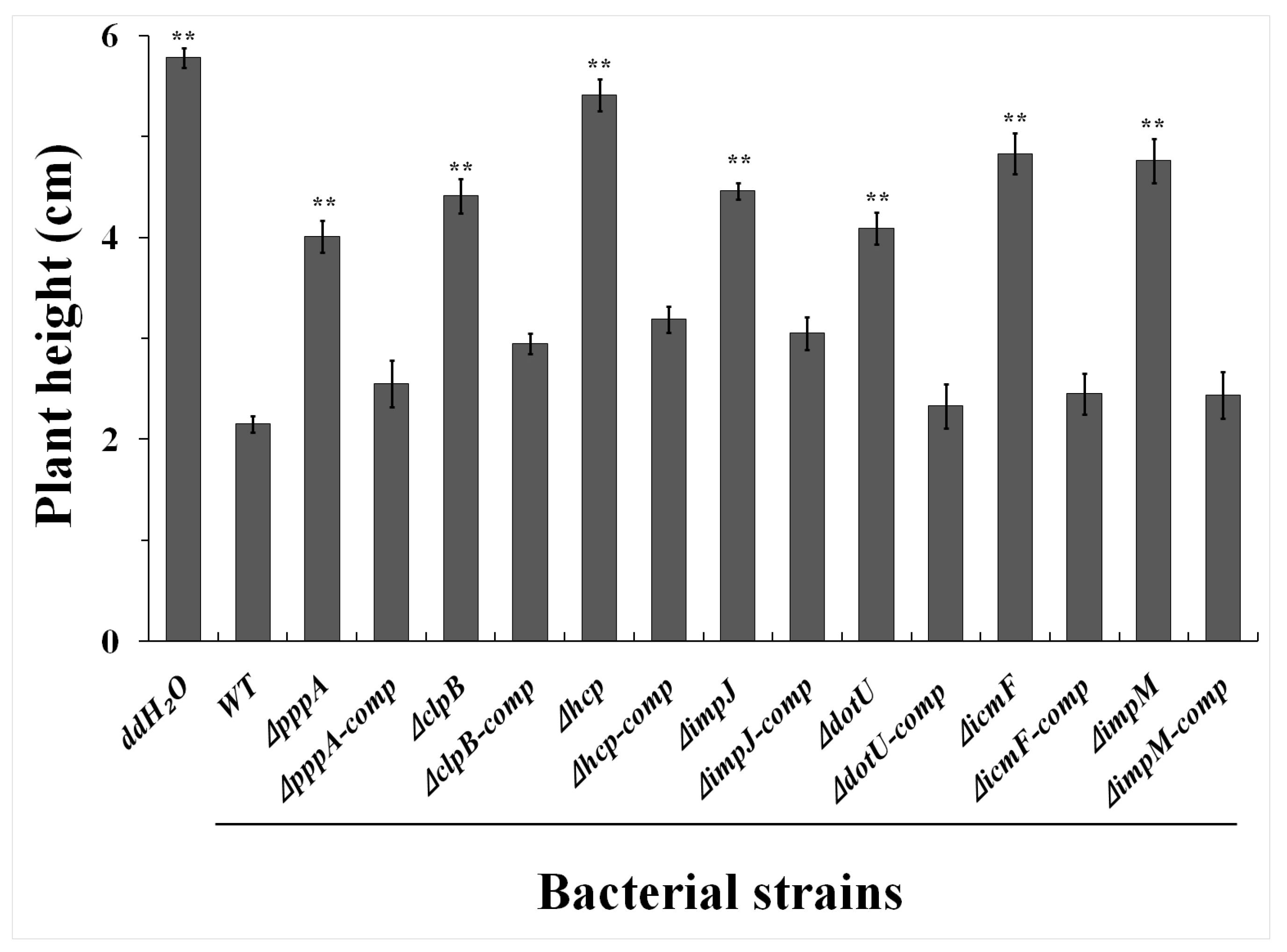
2.3. T6SS Affected Pathogenicity of Aaa Strain RS2 to Rice Seedlings
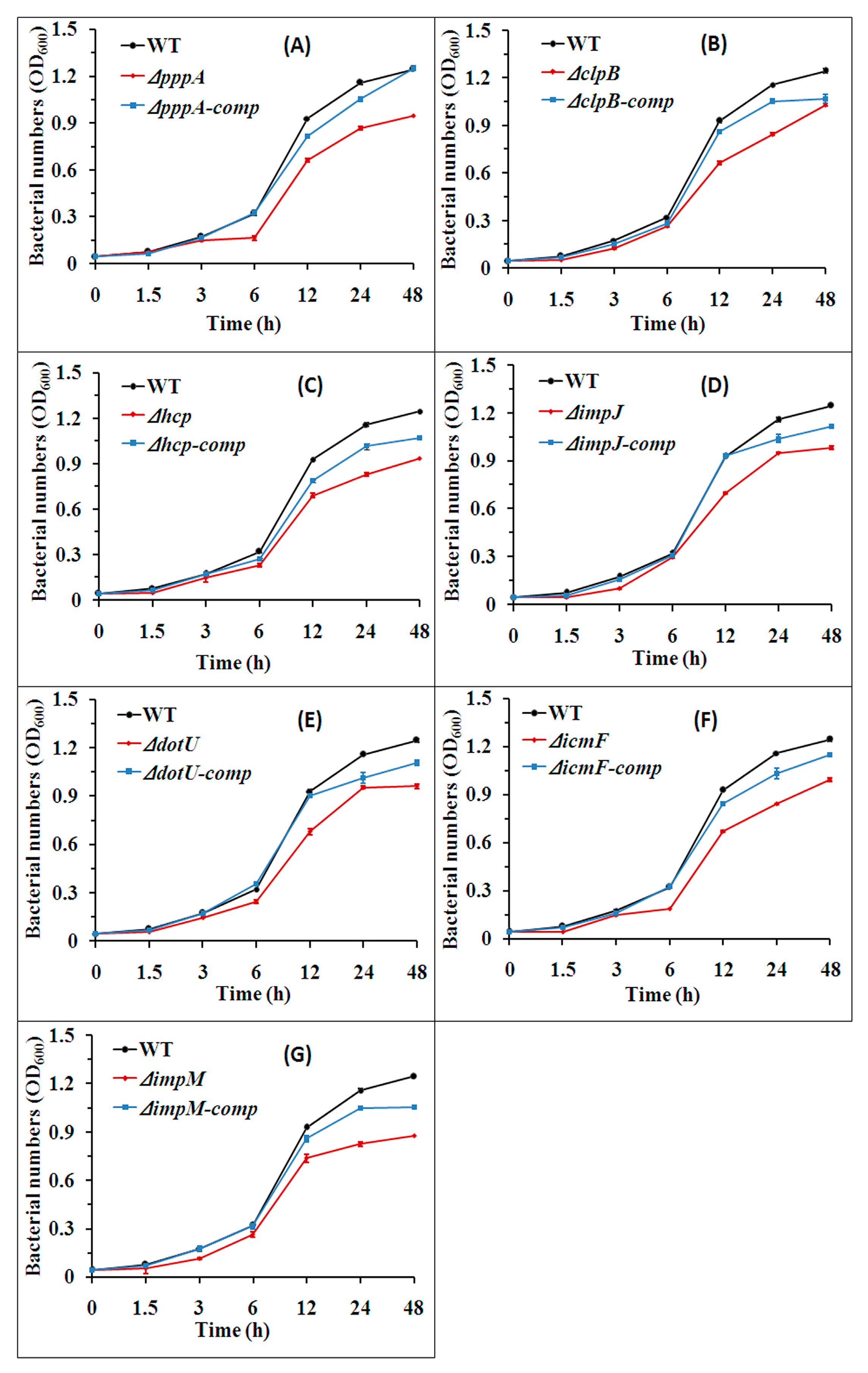
2.4. T6SS Affected the Growth of Aaa Strain RS2
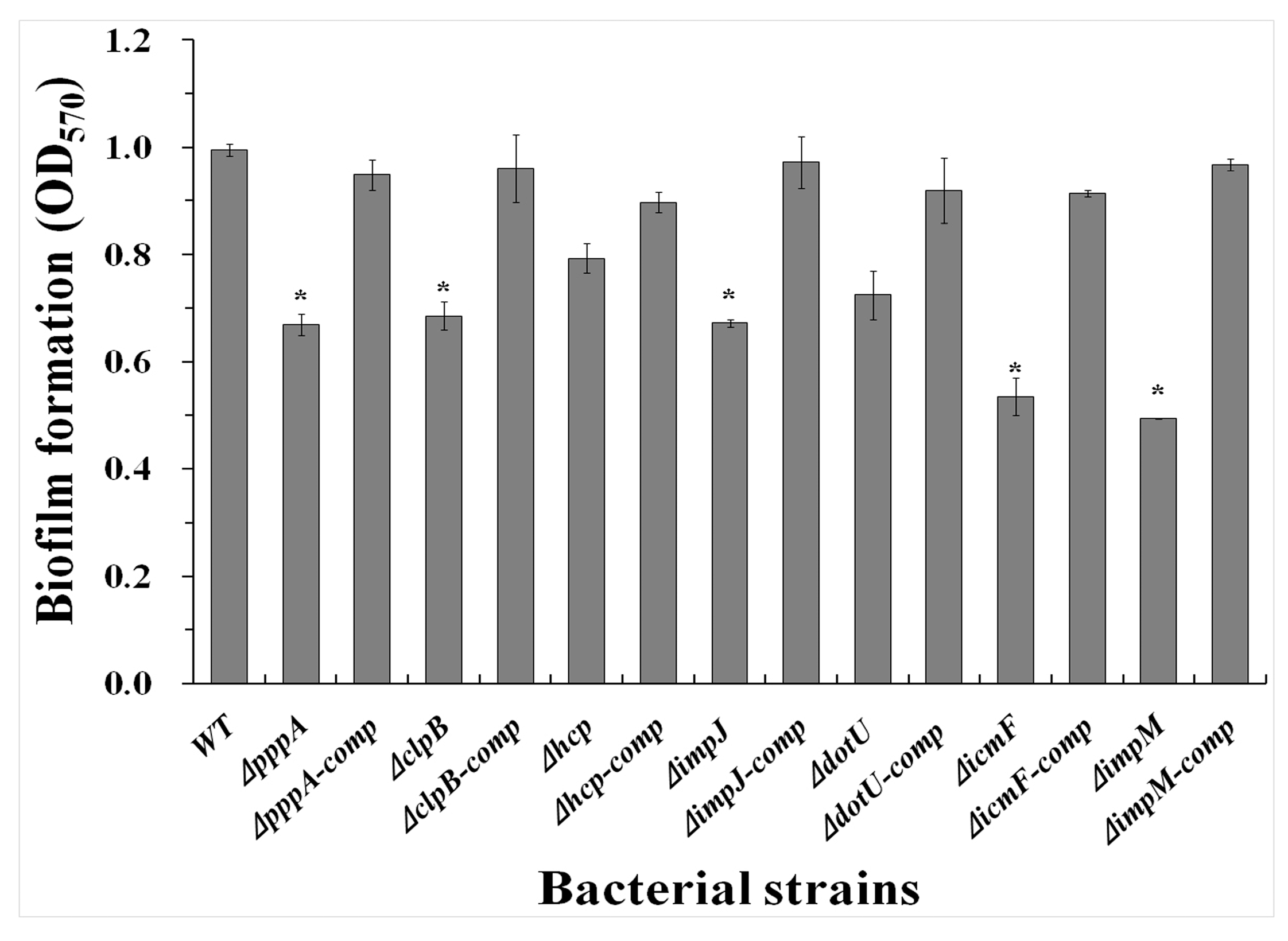
2.5. T6SS Affected the Biofilm Formation of Aaa Strain RS2
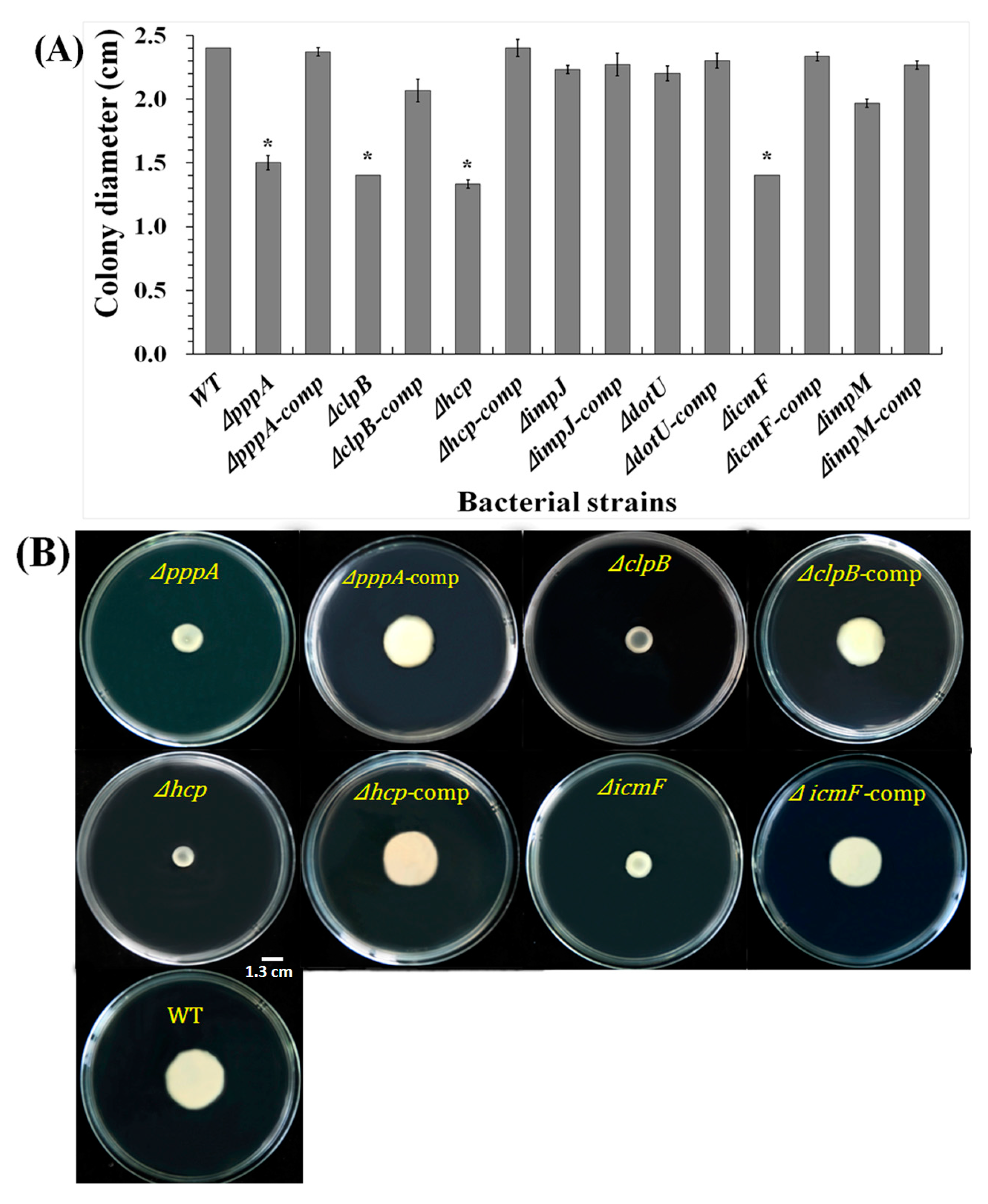
2.6. T6SS Affected the Swimming Motility of Aaa Strain RS2
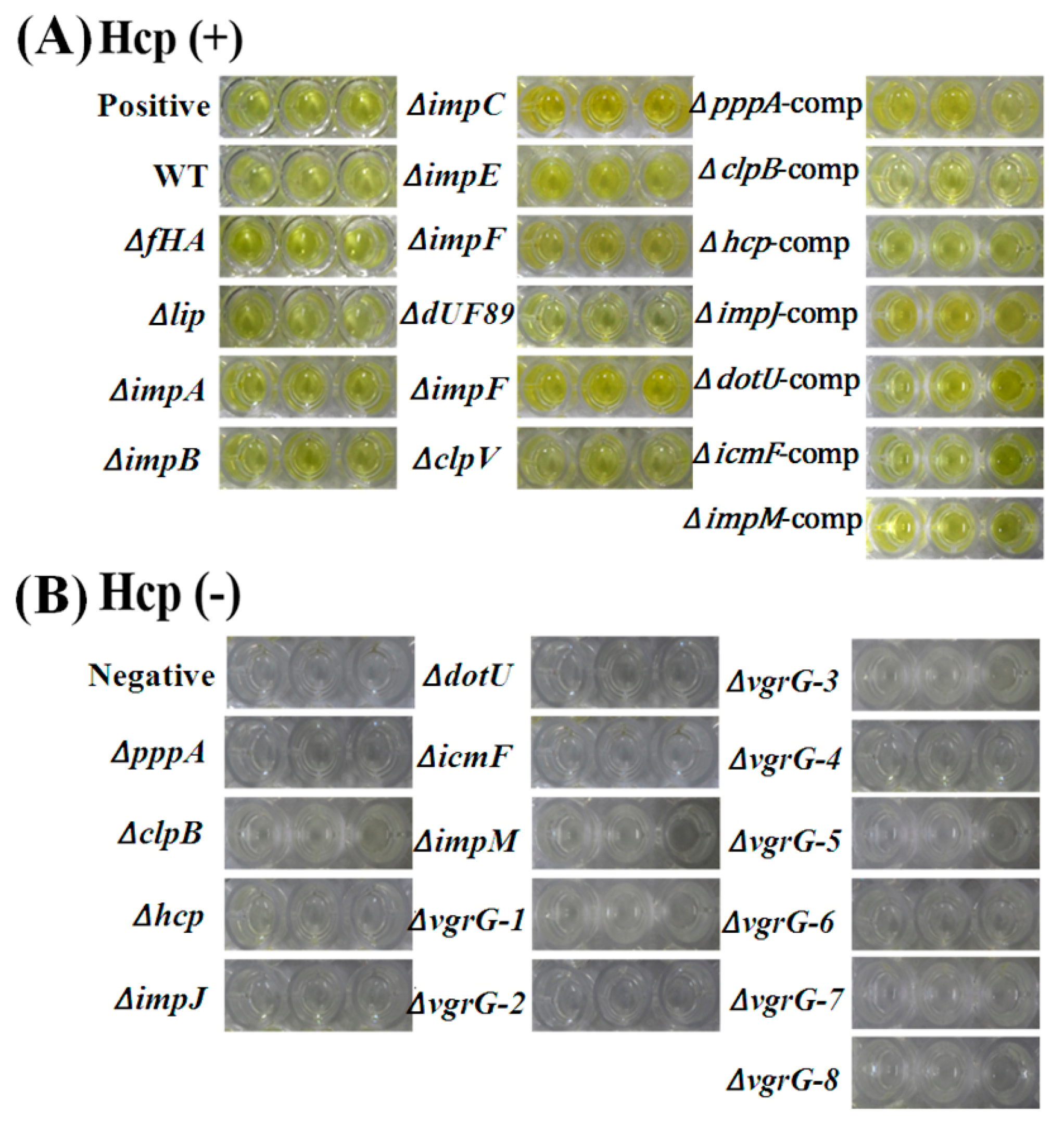
2.7. T6SS Affected Hcp Secretion of Aaa Strain RS2
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Growth Media and Inoculum Preparation
4.2. DNA Extraction and Amplification
4.3. In Silico Analysis of Type VI Secretion Loci
4.4. Generation of T6SS Mutants and Complementation
4.5. Seedling Pathogenicity in Rice
4.6. Bacterial Growth Assays
4.7. Biofilm Formation Measurement
4.8. Motility Assays
4.9. Measurement of the Secreted Hcp by ELISA
4.10. Statistical Analyses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Song, W.Y.; Kim, H.M.; Hwang, C.Y.; Schaad, N.W. Detection of Acidovorax avenae subsp. avenae in rice seeds using BIO-PCR. J. Phytopathol. 2004, 152, 667–676. [Google Scholar] [CrossRef]
- Shakya, D.D.; VInther, F.; Mathur, S.B. World wide distribution of a bacterial stripe pathogen of rice identified as Pseudomonas avenae. J. Phytopathol. 1985, 114, 256–259. [Google Scholar] [CrossRef]
- Cui, Z.; Ojaghian, M.R.; Tao, Z.; Kakar, K.U.; Zeng, J.; Zhao, W.; Duan, Y.; Vera Cruz, C.M.; Li, B.; Zhu, B.; et al. Multiplex pcr assay for simultaneous detection of six major bacterial pathogens of rice. J. Appl. Microbiol. 2016, 120, 1357–1367. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, Q.; Li, B.; Liu, B.; Wu, G.; Ibrahim, M.; Xie, G.; Li, H.; Sun, G. Differentiation in MALDI-TOF MS and FTIR spectra between two closely related species Acidovoraxoryzae and Acidovorax citrulli. BMC Microbiol. 2012, 12, 182. [Google Scholar] [CrossRef] [PubMed]
- Kadota, I.; Ohuchi, A.; Nishiyama, K. Serological properties and specificity of Pseudomonas avenae Manns 1909, the causal agent of bacterial brown stripe of rice. Jpn. J. Phytopathol. 1991, 57, 268–273. [Google Scholar] [CrossRef]
- Tian, Y.L.; Zhao, Y.Q.; Wu, X.R.; Liu, F.Q.; Hu, B.S.; Walcott, R.R. The type VI protein secretion system contributes to biofilm formation and seed-to-seedling transmission of Acidovorax citrulli on melon. Mol. Plant Pathol. 2015, 16, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.-L.; Zhang, G.-Q.; Liu, H.; Lou, M.-M.; Tian, W.-X.; Li, B.; Zhou, X.-P.; Zhu, B.; Jin, G.-L. Genome sequence of the rice-pathogenic bacterium Acidovorax avenae subsp. avenae RS-1. J. Bacteriol. 2011, 193, 5013–5014. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, B.; Ge, M.; Zhou, K.; Wang, Y.; Luo, J.; Ibrahim, M.; Xie, G.; Sun, G. Inhibitory effect and mode of action of chitosan solution against rice bacterial brown stripe pathogen Acidovorax avenae subsp. avenae RS-1. Carbohydr. Res. 2014, 391, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liu, B.; Yu, R.; Tao, Z.; Wang, Y.; Xie, G.; Li, H.; Sun, G. Bacterial brown stripe of rice in soil-less culture system caused by Acidovorax avenae subsp. avenae in China. J. Gen. Plant Pathol. 2011, 77, 64–67. [Google Scholar] [CrossRef]
- Li, B.; Wang, L.; Ibrahim, M.; Ge, M.; Wang, Y.; Mannan, S.; Asif, M.; Sun, G. Membrane protein profiling of Acidovorax avenae subsp. avenaeunder various growth conditions. Arch. Microbiol. 2015, 197, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Ge, M.; Zhang, Y.; Wang, L.; Ibrahim, M.; Wang, Y.; Sun, G.; Chen, G. New insights into virulence mechanisms of rice pathogen Acidovorax avenae subsp. avenaestrain RS-1 following exposure to β-lactam antibiotics. Sci. Rep. 2016, 6, 22241. [Google Scholar] [CrossRef] [PubMed]
- Mougous, J.D.; Cuff, M.E.; Raunser, S.; Shen, A.; Zhou, M.; Gifford, C.A.; Goodman, A.L.; Joachimiak, G.; Ordoñez, C.L.; Lory, S.; et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 2006, 312, 1526–1530. [Google Scholar] [CrossRef] [PubMed]
- Costa, T.R.D.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion systems in gram-negative bacteria: Structural and mechanistic insights. Nat. Rev. Microl. 2015, 13, 343–359. [Google Scholar] [CrossRef] [PubMed]
- Pukatzki, S.; Ma, A.T.; Sturtevant, D.; Krastins, B.; Sarracino, D.; Nelson, W.C.; Heidelberg, J.F.; Mekalanos, J.J. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 2006, 103, 1528–1533. [Google Scholar] [CrossRef] [PubMed]
- Leiman, P.G.; Basler, M.; Ramagopal, U.A.; Bonanno, J.B.; Sauder, J.M.; Pukatzki, S.; Burley, S.K.; Almo, S.C.; Mekalanos, J.J. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. USA 2009, 106, 4154–4159. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gu, D.; Sheng, L.; Wang, Q.; Zhang, Y. Investigation of the roles of T6SS genes in motility, biofilm formation, and extracellular protease asp production in Vibrio alginolyticus with modified gateway-compatible plasmids. Lett. Appl. Microbiol. 2012, 55, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Miyata, S.T.; Kitaoka, M.; Brooks, T.M.; McAuley, S.B.; Pukatzki, S. Vibrio cholerae requires the type VI secretion system virulence factor Vasx to kill Dictyostelium discoideum. Infect. Immun. 2011, 79, 2941–2949. [Google Scholar] [CrossRef] [PubMed]
- Weber, B.; Hasic, M.; Chen, C.; Wai, S.N.; Milton, D.L. Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ. Microbiol. 2009, 11, 3018–3028. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, F.; Li, B.; Yang, Y.Z.; Ibrahim, M.; Fang, Y.S.; Qiu, W.; Masum, M.M.I.; Oliva, R. Characterization and functional analysis of clpB gene from Acidovorax avenae subsp. avenae RS-1. Plant Pathol. 2017. [Google Scholar] [CrossRef]
- Bingle, L.E.; Bailey, C.M.; Pallen, M.J. Type VI secretion: A beginner’s guide. Curr. Opin. Microbiol. 2008, 11, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Boyer, F.; Fichant, G.; Berthod, J.; Vandenbrouck, Y.; Attree, I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: What can be learned from available microbial genomic resources? BMC Genom. 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Schell, M.A.; Ulrich, R.L.; Ribot, W.J.; Brueggemann, E.E.; Hines, H.B.; Chen, D.; Lipscomb, L.; Kim, H.S.; Mrázek, J.; Nierman, W.C.; et al. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 2007, 64, 1466–1485. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S.; Mande, S.S. Identification and functional characterization of gene components of type VI secretion system in bacterial genomes. PLoS ONE 2008, 3, e2955. [Google Scholar] [CrossRef] [PubMed]
- Pukatzki, S.; McAuley, S.B.; Miyata, S.T. The type VI secretion system: Translocation of effectors and effector-domains. Curr. Opin. Microbiol. 2009, 12, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Tao, J.; Yu, H.; Ni, J.J.; Zeng, L.B.; Teng, Q.H.; Kim, K.S.; Zhao, G.P.; Guo, X.; Yao, Y. Hcp family proteins secreted via the type VI secretion system coordinately regulate Escherichia coli K1 interaction with human brain microvascular endothelial cells. Infect. Immun. 2012, 80, 1243–1251. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.; Shi, Y.; Qiu, H.; Li, B.; Jabeen, A.; Li, L.P.; Liu, H.; Kube, M.; Xie, G.; Wang, Y.; et al. Differential expression of in vivoand in vitro protein profile of outer membrane of Acidovorax avenae subsp. avenae. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Burtnick, M.N.; Brett, P.J.; Harding, S.V.; Ngugi, S.A.; Ribot, W.J.; Chantratita, N.; Scorpio, A.; Milne, T.S.; Dean, R.E.; Fritz, D.L.; et al. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 2011, 79, 1512–1525. [Google Scholar] [CrossRef] [PubMed]
- Alteri, C.J.; Mobley, H.L. The versatile type VI secretion system. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Li, B.; Ibrahim, M.; Ge, M.; Cui, Z.; Sun, G.; Xu, F.; Kube, M. Transcriptome analysis of Acidovorax avenae subsp. avenae cultivated in vivo and co-culture with Burkholderia seminalis. Sci. Rep. 2014, 4, 5698. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Jin, G.; Li, B.; Kakar, K.; Ojaghian, M.; Wang, Y.; Xie, G.; Sun, G. Gene expression of type VI secretion system associated with environmental survival in Acidovorax avenae subsp. avenaeby principle component analysis. Int. J. Mol. Sci. 2015, 16, 22008–22026. [Google Scholar] [CrossRef] [PubMed]
- Sheng, L.; Lv, Y.; Liu, Q.; Wang, Q.; Zhang, Y. Connecting type VI secretion, quorum sensing, and c-di-GMP production in fish pathogen Vibrio alginolyticus through phosphatase Pppa. Vet. Microbiol. 2013, 162, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.Y.; Siame, B.A.; Snowball, H.; Mok, Y.-K. Type VI secretion regulation: Cross talk and intracellular communication. Curr. Opin. Microbiol. 2011, 14, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Zoued, A.; Durand, E.; Bebeacua, C.; Brunet, Y.R.; Douzi, B.; Cambillau, C.; Cascales, E.; Journet, L. Tssk is a trimeric cytoplasmic protein interacting with components of both phage-like and membrane anchoring complexes of the type VI secretion system. J. Biol. Chem. 2013, 288, 27031–27041. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Waldor, M.K.; Mekalanos, J.J. Tn-seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 2013, 14, 652–663. [Google Scholar] [CrossRef] [PubMed]
- Suarez, G.; Sierra, J.C.; Kirtley, M.L.; Chopra, A.K. Role of Hcp, a type 6 secretion system effector, of Aeromonas hydrophila in modulating activation of host immune cells. Microbiology 2010, 156 Pt 12, 3678–3688. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Y.; Chung, P.C.; Shih, H.W.; Wen, S.R.; Lai, E.M. Secretome analysis uncovers an Hcp-family protein secreted via a type VI secretion system in Agrobacterium tumefaciens. J. Bacteriol. 2008, 190, 2841–2850. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.Y.; Hao, S.; Lan, R.T.; Wang, G.X.; Xiao, D.; Sun, H.; Xu, J. The type VI secretion system modulates flagellar gene expression and secretion in Citrobacter freundii and contributes to adhesion and cytotoxicity to host cells. Infect. Immun. 2015, 83, 2596–2604. [Google Scholar] [CrossRef] [PubMed]
- Castiblanco, L.F.; Sundin, G.W. New insights on molecular regulation of biofilm formation in plant-associated bacteria. J. Integr. Plant Biol. 2016, 58, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Bogino, P.C.; Oliva Mde, L.; Sorroche, F.G.; Giordano, W. The role of bacterial biofilms and surface components in plant-bacterial associations. Int. J. Mol. Sci. 2013, 14, 15838–15859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallique, M.; Decoin, V.; Barbey, C.; Rosay, T.; Feuilloley, M.G.J.; Orange, N.; Merieau, A. Contribution of the Pseudomonas fluorescens MFE01 type VI secretion system to biofilm formation. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Mizan, M.F.R.; Jahid, I.K.; Kim, M.; Lee, K.H.; Kim, T.J.; Ha, S.D. Variability in biofilm formation correlates with hydrophobicity and quorum sensing among Vibrio parahaemolyticus isolates from food contact surfaces and the distribution of the genes involved in biofilm formation. Biofouling 2016, 32, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning:A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Penfold, R.J.; Pemberton, J.M. An Improved Suicide Vector for Construction of Chromosomal Insertion Mutations in Bacteria; Elsevier: Amsterdam, The Netherlands, 1992. [Google Scholar]
- Simon, R.; Priefer, U.; Puhler, A. A broad host range mobilization system for in vivogenetic engineering: Transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Gao, G.; Lu, H.; Huang, L.; Hua, Y. Construction of DNA damage response gene pprI function-deficient and function-complementary mutants in Deinococcus radiodurans. Chin. Sci. Bull. 2005, 50, 311–316. [Google Scholar] [CrossRef]
- Smith, M.D.; Guild, W.R. Improved method for conjugative transfer by filter mating of Streptococcus pneumoniae. J. Bacteriol. 1980, 144, 457–459. [Google Scholar] [PubMed]
- Peeters, E.; Nelis, H.J.; Coenye, T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 2008, 72, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Tian, W.-X.; Ibrahim, M.; Li, B.; Zhang, G.-Q.; Zhu, B.; Xie, G.-L. Chacterization of pilP, a gene required for twitching motility, pathogenicity, and biofilm formation of Acidovoraxavenaesubsp. Avenae RS-1. Eur. J. Plant Pathol. 2012, 134, 551–560. [Google Scholar] [CrossRef]
- Slutzki, M.; Barak, Y.; Reshef, D.; Schueler-Furman, O.; Lamed, R.; Bayer, E.A. Indirect elisa-based approach for comparative measurement of high-affinity cohesin-dockerin interactions. J. Mol. Recognit. 2012, 25, 616–622. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masum, M.M.I.; Yang, Y.; Li, B.; Olaitan, O.S.; Chen, J.; Zhang, Y.; Fang, Y.; Qiu, W.; Wang, Y.; Sun, G. Role of the Genes of Type VI Secretion System in Virulence of Rice Bacterial Brown Stripe Pathogen Acidovorax avenae subsp. avenae Strain RS-2. Int. J. Mol. Sci. 2017, 18, 2024. https://doi.org/10.3390/ijms18102024
Masum MMI, Yang Y, Li B, Olaitan OS, Chen J, Zhang Y, Fang Y, Qiu W, Wang Y, Sun G. Role of the Genes of Type VI Secretion System in Virulence of Rice Bacterial Brown Stripe Pathogen Acidovorax avenae subsp. avenae Strain RS-2. International Journal of Molecular Sciences. 2017; 18(10):2024. https://doi.org/10.3390/ijms18102024
Chicago/Turabian StyleMasum, Md. Mahidul Islam, Yingzi Yang, Bin Li, Ogunyemi Solabomi Olaitan, Jie Chen, Yang Zhang, Yushi Fang, Wen Qiu, Yanli Wang, and Guochang Sun. 2017. "Role of the Genes of Type VI Secretion System in Virulence of Rice Bacterial Brown Stripe Pathogen Acidovorax avenae subsp. avenae Strain RS-2" International Journal of Molecular Sciences 18, no. 10: 2024. https://doi.org/10.3390/ijms18102024





