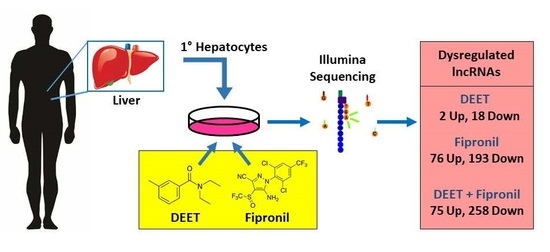
Differential Expression Profile of lncRNAs from Primary Human Hepatocytes Following DEET and Fipronil Exposure
Abstract
:1. Introduction
2. Results and Discussion
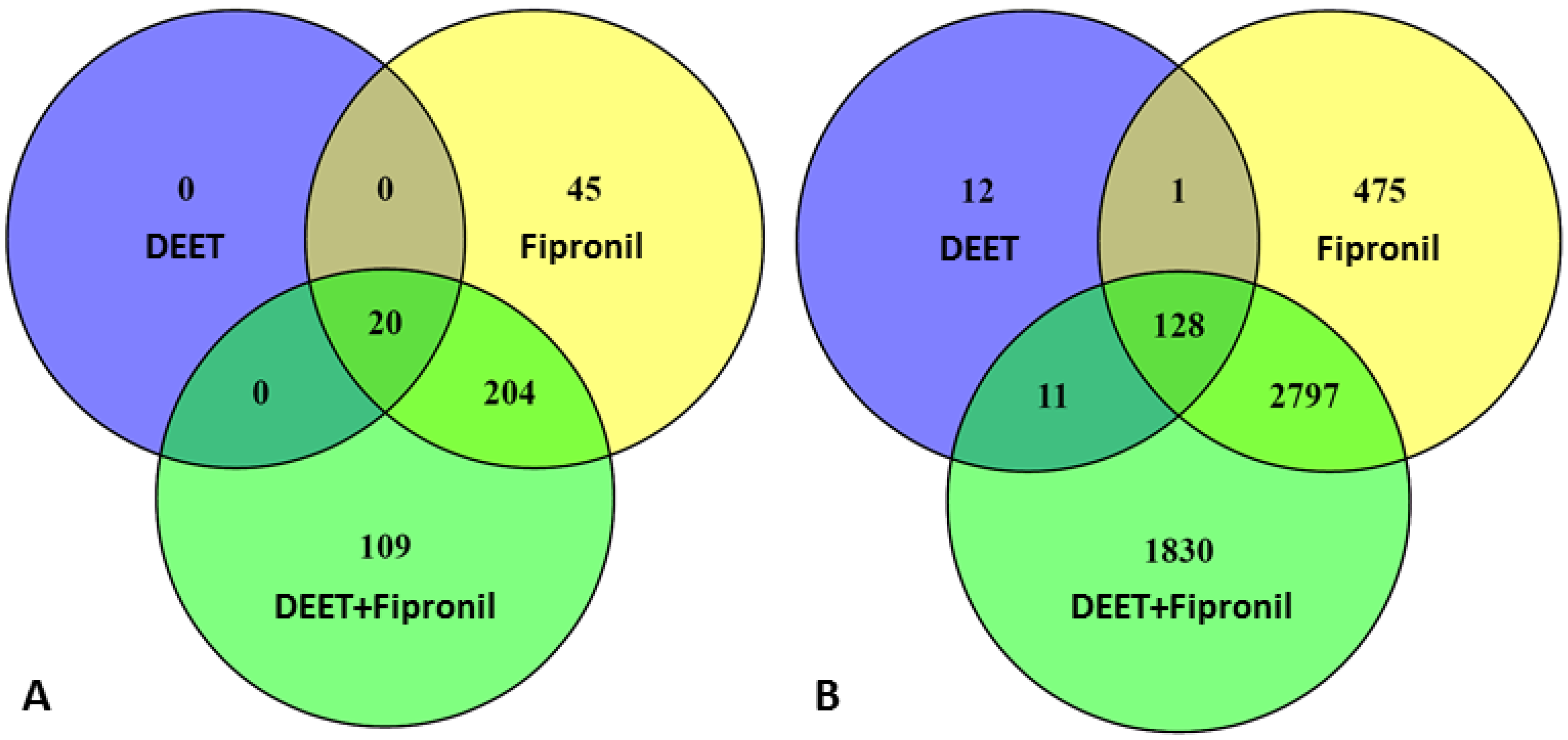
2.1. Effects of DEET and Fipronil on LncRNA Versus Protein-Coding Gene Transcription
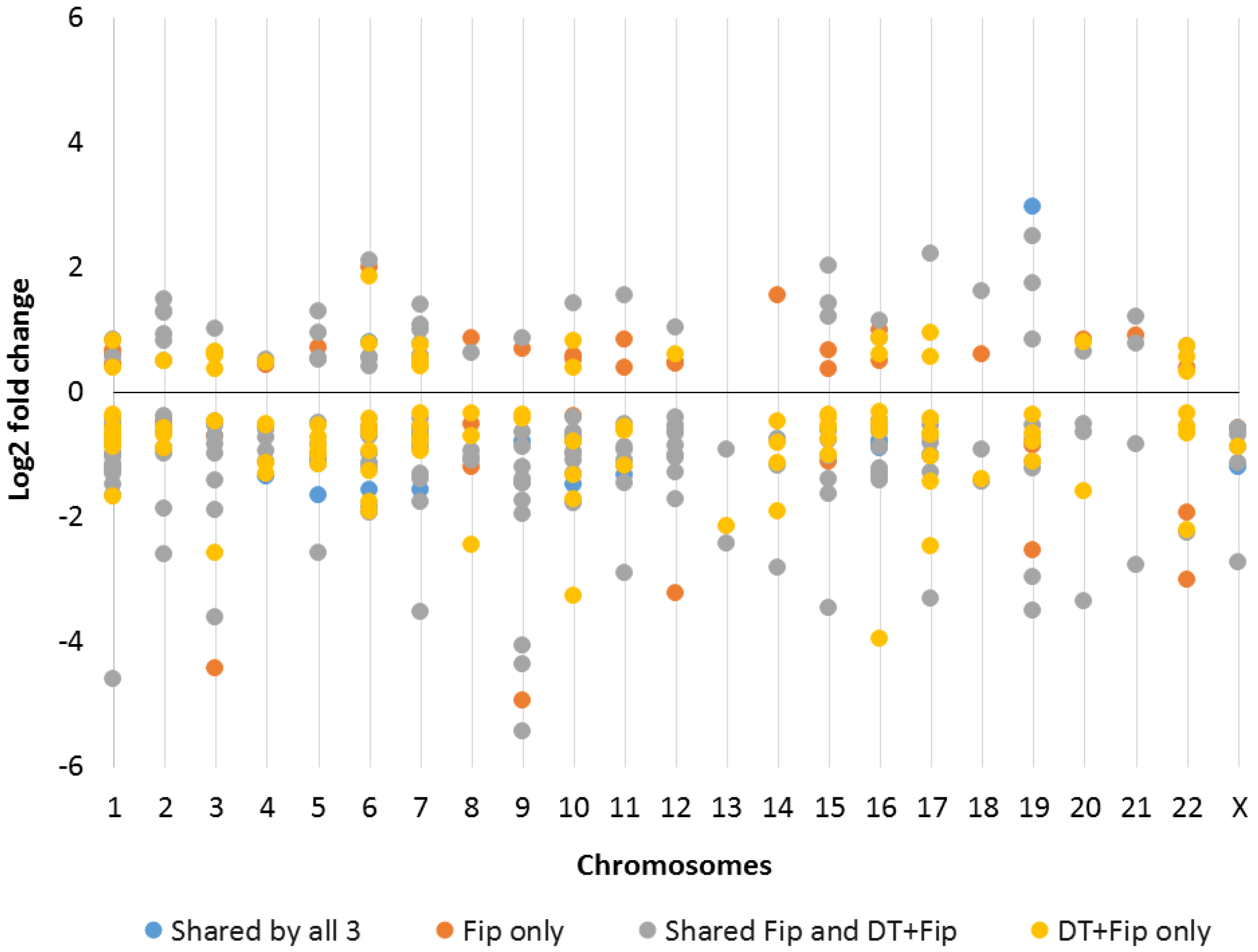
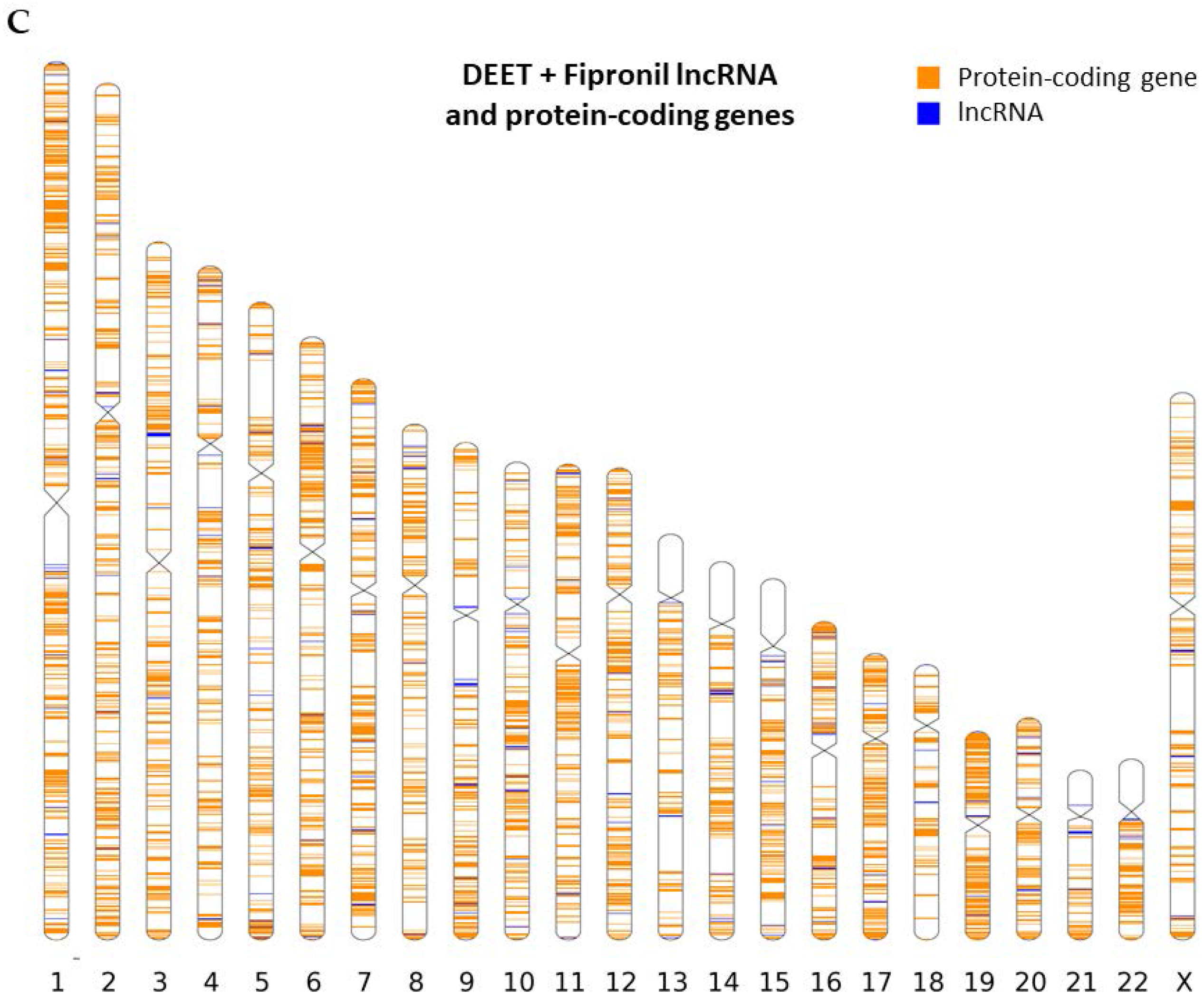
2.2. Chromosomal Mapping of Dysregulated LncRNAs
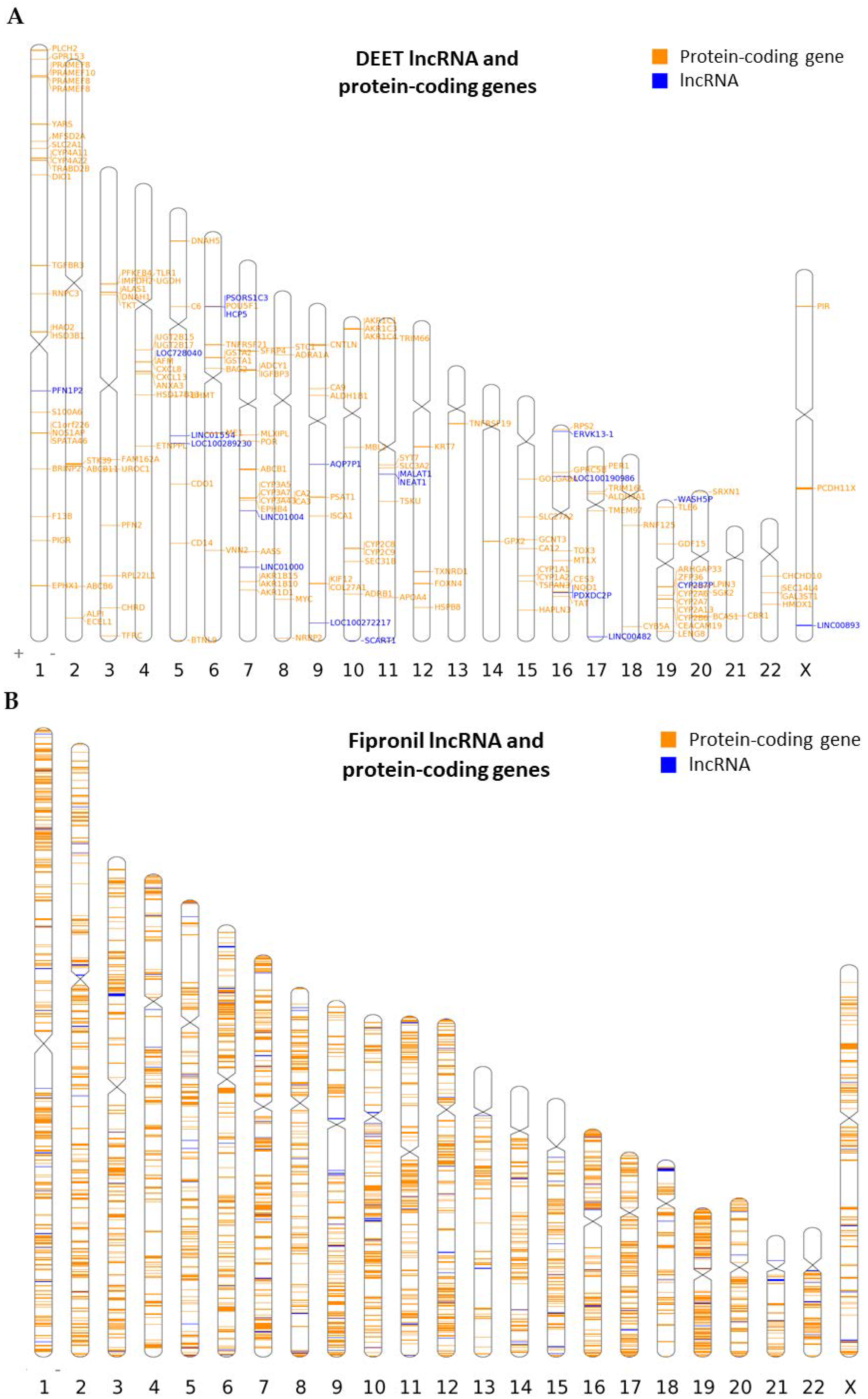
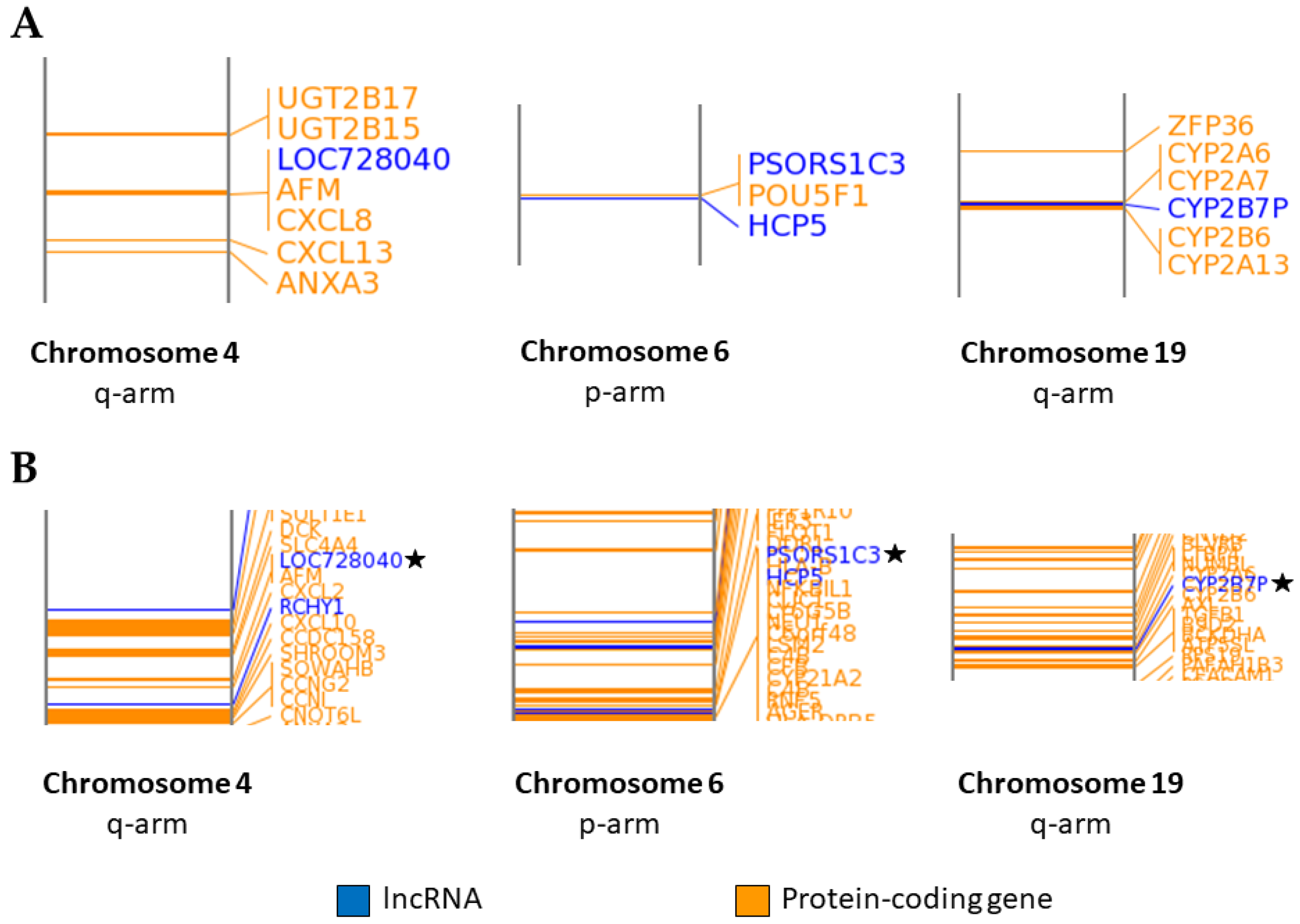
2.3. Association of LncRNA Genes with Protein-Coding Genes
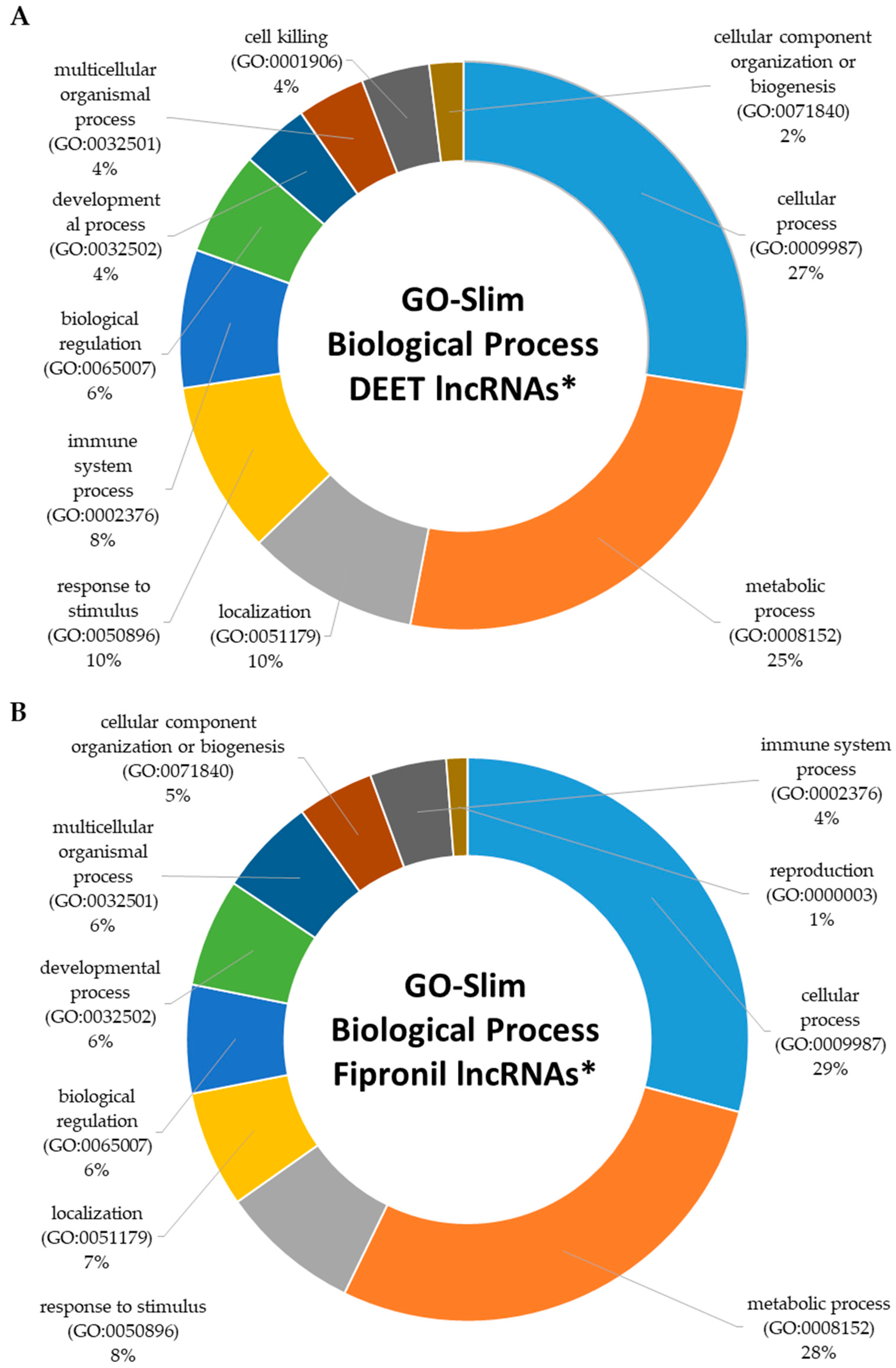
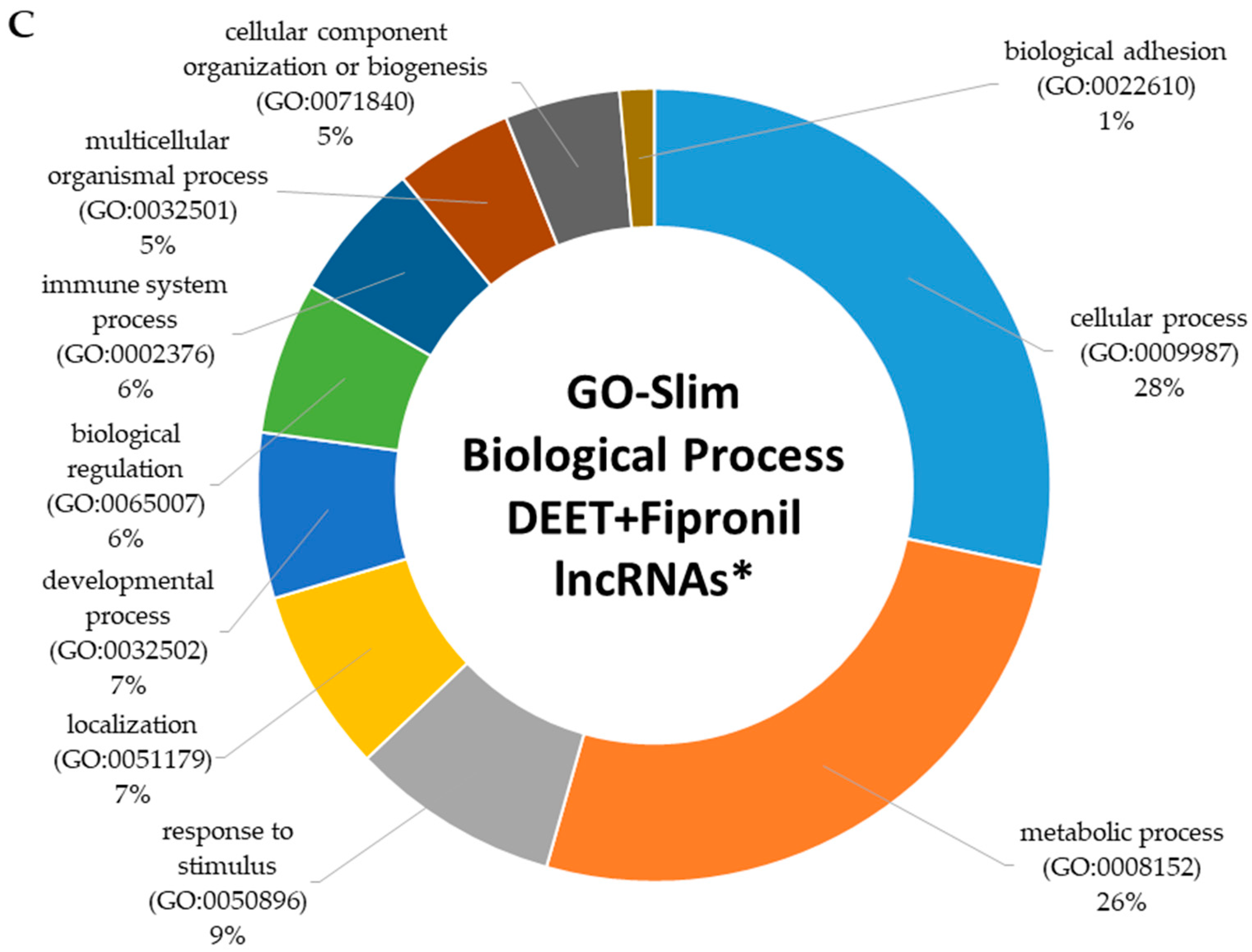
2.4. Functional Characterization of Dysregulated LncRNAs
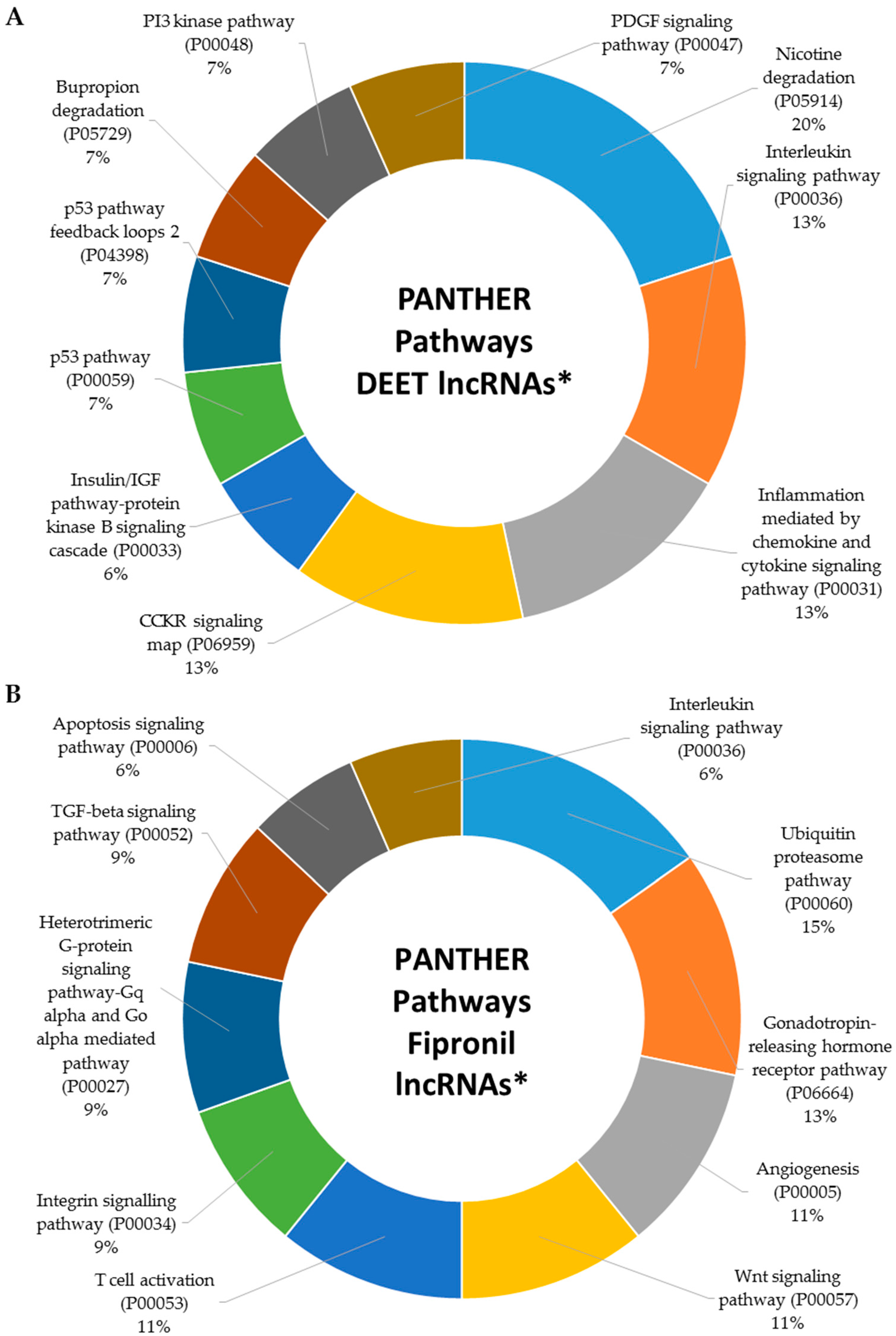
2.5. Metabolic Signaling Pathways Associated with Dysregulated LncRNAs
2.5.1. Dysregulated LncRNAs Involved in Innate and Adaptive Immunity
2.5.2. Dysregulated LncRNAs Involved in the Transformation-Related Protein 53 (p53) Signaling Pathway
2.6. Specific Well-Studied LncRNAs Dysregulated by DEET and Fipronil
3. Materials and Methods
3.1. Primary Human Hepatocytes
3.2. DEET and Fipronil Treatments
3.3. RNA Isolation and Quality Assessment
3.4. Illumina Sequencing
3.5. RNA-Seq Analysis
3.6. Dysregulated LncRNA Characterization
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| DEET | N,N-diethyl-m-toluamide |
| NCBI | National Center for Biotechnology Information |
| TSS | Transcription start site |
| HUGO | Human Genome Organisation |
| HGNC | HUGO Gene Nomenclature Committee |
| GREAT | Genomic regions enrichment of annotations tool |
| PANTHER | Protein analysis through evolutionary relationships |
References
- Bernhardt, E.S.; Rosi, E.J.; Gessner, M.O. Synthetic chemicals as agents of global change. Front. Ecol. Environ. 2017, 15, 84–90. [Google Scholar] [CrossRef]
- Veltri, J.C.; Osimitz, T.G.; Bradford, D.C.; Page, B.C. Retrospective analysis of calls to poison control centers resulting from exposure to the insect repellent N,N-diethyl-m-toluamide (deet) from 1985–1989. J. Toxicol. Clin. Toxicol. 1994, 32, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Mlakar, J.; Korva, M.; Tul, N.; Popović, M.; Poljšak-Prijatelj, M.; Mraz, J.; Kolenc, M.; Resman Rus, K.; Vesnaver Vipotnik, T.; Fabjan Vodušek, V. Zika virus associated with microcephaly. N. Engl. J. Med. 2016, 2016, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Morris, H. Zika: The Latest Advice for Travellers. Available online: http://www.telegraph.co.uk/travel/news/Zika-virus-medical-advice-for-travellers/ (accessed on 22 February 2017).
- Vargo, E.L.; Parman, V. Effect of fipronil on subterranean termite colonies (isoptera: Rhinotermitidae) in the field. J. Econ. Entomol. 2012, 105, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Carrington, D. Eu to Ban Fipronil to Protect Honeybees. Available online: https://www.theguardian.com/environment/2013/jul/16/eu-fipronil-ban-bees (accessed on 22 February 2017).
- Hamon, N.; Gamboa, H.; Ernesto, J.; Garcia, M. Fipronil: A major advance for the control of boll weevil in colombia. In Proceedings of the Beltwide Cotton Conferences, Nashville, TN, USA, 9–12 January 1996. [Google Scholar]
- Tingle, C.C.; Rother, J.A.; Dewhurst, C.F.; Lauer, S.; King, W.J. Fipronil: Environmental fate, ecotoxicology, and human health concerns. In Reviews of Environmental Contamination and Toxicology; Ware, G.W., Ed.; Springer: New York, NY, USA, 2003; pp. 1–66. ISBN 978-1-4899-7283-5. [Google Scholar]
- Mohamed, F.; Senarathna, L.; Percy, A.; Abeyewardene, M.; Eaglesham, G.; Cheng, R.; Azher, S.; Hittarage, A.; Dissanayake, W.; Sheriff, M.R. Acute human self-poisoning with the N-phenylpyrazole insecticide fipronil—A gabaa-gated chloride channel blocker. J. Toxicol. Clin. Toxicol. 2004, 42, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Schoenig, G.P.; Osimitz, T.G.; Gabriel, K.L.; Hartnagel, R.; Gill, M.W.; Goldenthal, E.I. Evaluation of the chronic toxicity and oncogenicity of N,N-diethyl-m-toluamide (deet). Toxicol. Sci. 1999, 47, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Heffernan, A.; English, K.; Toms, L.; Calafat, A.; Valentin-Blasini, L.; Hobson, P.; Broomhall, S.; Ware, R.; Jagals, P.; Sly, P. Cross-sectional biomonitoring study of pesticide exposures in queensland, australia, using pooled urine samples. Environ. Sci. Pollut. Res. 2016, 23, 23436–23448. [Google Scholar] [CrossRef] [PubMed]
- Herin, F.; Boutet-Robinet, E.; Levant, A.; Dulaurent, S.; Manika, M.; Galatry-Bouju, F.; Caron, P.; Soulat, J.-M. Thyroid function tests in persons with occupational exposure to fipronil. Thyroid 2011, 21, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Usmani, K.A.; Rose, R.L.; Goldstein, J.A.; Taylor, W.G.; Brimfield, A.A.; Hodgson, E. In vitro human metabolism and interactions of repellent N,N-diethyl-m-toluamide. Drug Metab. Dispos. 2002, 30, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Selim, S.; Hartnagel, R.E.; Osimitz, T.G.; Gabriel, K.L.; Schoenig, G.P. Absorption, metabolism, and excretion of N,N-diethyl-m-toluamide following dermal application to human volunteers. Fundam. Appl. Toxicol. 1995, 25, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Das, P.C.; Cao, Y.; Cherrington, N.; Hodgson, E.; Rose, R.L. Fipronil induces cyp isoforms and cytotoxicity in human hepatocytes. Chem. Biol. Interact. 2006, 164, 200–214. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Usmani, K.A.; Hodgson, E.; Rose, R.L. In vitro metabolism of fipronil by human and rat cytochrome p450 and its interactions with testosterone and diazepam. Chem. Biol. Interact. 2004, 147, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Toxicology Data Network. Fipronil. Available online: https://toxnet.nlm.nih.gov/cgi-bin/sis/search/a?dbs+hsdb:@term+@DOCNO+7051 (accessed on 10 April 2017).
- Mitchell, R.D.; Dhammi, A.; Wallace, A.; Hodgson, E.; Roe, R.M. Impact of environmental chemicals on the transcriptome of primary human hepatocytes: Potential for health effects. J. Biochem. Mol. Toxicol. 2016, 30, 375–395. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cao, X. Long noncoding RNAs in innate immunity. Cell. Mol. Immunol. 2016, 13, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Xu, M.; Mo, Y.-Y. Role of the lncRNA–p53 regulatory network in cancer. J. Mol. Cell Biol. 2014, 6, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Villegas, V.E.; Zaphiropoulos, P.G. Neighboring gene regulation by antisense long non-coding RNAs. Int. J. Mol. Sci. 2015, 16, 3251–3266. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.T.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Aken, B.L.; Ayling, S.; Barrell, D.; Clarke, L.; Curwen, V.; Fairley, S.; Banet, J.F.; Billis, K.; Girón, C.G.; Hourlier, T. The ensembl gene annotation system. Database 2016, 2016, baw093. [Google Scholar] [CrossRef] [PubMed]
- Milligan, M.J.; Lipovich, L. Pseudogene-derived lncRNAs: Emerging regulators of gene expression. Front. Genet. 2015, 5, 476. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.M.; Zheng, D.; Zhang, Z.; Carriero, N.; Gerstein, M. Transcribed processed pseudogenes in the human genome: An intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res. 2005, 33, 2374–2383. [Google Scholar] [CrossRef] [PubMed]
- Milligan, M.J.; Harvey, E.; Yu, A.; Morgan, A.L.; Smith, D.L.; Zhang, E.; Berengut, J.; Sivananthan, J.; Subramaniam, R.; Skoric, A. Global intersection of long non-coding RNAs with processed and unprocessed pseudogenes in the human genome. Front. Genet. 2016, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Tam, O.H.; Aravin, A.A.; Stein, P.; Girard, A.; Murchison, E.P.; Cheloufi, S.; Hodges, E.; Anger, M.; Sachidanandam, R.; Schultz, R.M. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 2008, 453, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Cheng, E.-C.; Zhong, M.; Lin, H. Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res. 2015, 25, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef] [PubMed]
- NCBI, R.C. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2017, 45, D12. [Google Scholar]
- Branca, R.M.; Orre, L.M.; Johansson, H.J.; Granholm, V.; Huss, M.; Pérez-Bercoff, Å.; Forshed, J.; Käll, L.; Lehtiö, J. Hirief lc-ms enables deep proteome coverage and unbiased proteogenomics. Nat. Methods 2014, 11, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Dündar, F.; Skrabanek, L.; Zumbo, P. Introduction to differential gene expression analysis using RNA-seq. Appl. Bioinformatics 2015, 1–67. [Google Scholar]
- Gray, K.A.; Yates, B.; Seal, R.L.; Wright, M.W.; Bruford, E.A. Genenames. org: The HGNC resources in 2015. Nucleic Acids Res. 2014, 43, 1079–1085. [Google Scholar] [CrossRef] [PubMed]
- Kin, T.; Ono, Y. Idiographica: A general-purpose web application to build idiograms on-demand for human, mouse and rat. Bioinformatics 2007, 23, 2945–2946. [Google Scholar] [CrossRef] [PubMed]
- McLean, C.Y.; Bristor, D.; Hiller, M.; Clarke, S.L.; Schaar, B.T.; Lowe, C.B.; Wenger, A.M.; Bejerano, G. Great improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010, 28, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Stamatoyannopoulos, J.A. Illuminating eukaryotic transcription start sites. Nat. Methods 2010, 7, 501–503. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2016, 45, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Ganeshan, K.; Chawla, A. Metabolic regulation of immune responses. Annu. Rev. Immunol. 2014, 32, 609–634. [Google Scholar] [CrossRef] [PubMed]
- Tomar, N.; De, R.K. Cross talk between the metabolic and immune systems. Immunoinformatics 2014, 1184, 13–21. [Google Scholar]
- Puzio-Kuter, A.M. The role of p53 in metabolic regulation. Genes Cancer 2011, 2, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-D.; Qin, Z.-H.; Wang, J. The role of p53 in cell metabolism. Acta Pharmacol. Sin. 2010, 31, 1208. [Google Scholar] [CrossRef] [PubMed]
- WHO. U.N. Issues List of 12 Most Worrying Drug-Resistant Bacteria. Available online: http://www.cbsnews.com/news/un-list-12-most-worrying-bacteria-antibiotic-resistant-superbugs/ (accessed on 27 February 2017).
- Liu, Y.; Zhao, J.; Zhang, W.; Gan, J.; Hu, C.; Huang, G.; Zhang, Y. LncRNA gas5 enhances g1 cell cycle arrest via binding to ybx1 to regulate p21 expression in stomach cancer. Sci. Rep. 2015, 5, 10159. [Google Scholar] [CrossRef] [PubMed]
- Mourtada-Maarabouni, M.; Hedge, V.L.; Kirkham, L.; Farzaneh, F.; Williams, G.T. Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (gas5). J. Cell Sci. 2008, 121, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Van Manen, D.; Kootstra, N.A.; Boeser-Nunnink, B.; Handulle, M.A.; van’t Wout, A.B.; Schuitemaker, H. Association of HLA-c and HCP5 gene regions with the clinical course of HIV-1 infection. Aids 2009, 23, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W. Mutations of the braf gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Read, A.; Strachan, T. Chapter 18: Cancer genetics. Hum. Mol. Genet. 1999, 2. [Google Scholar]
- Harris, S.L.; Levine, A.J. The p53 pathway: Positive and negative feedback loops. Oncogene 2005, 24, 2899–2908. [Google Scholar] [CrossRef] [PubMed]
- Moll, U.M.; Petrenko, O. The mdm2-p53 interaction. Mol. Cancer Res. 2003, 1, 1001–1008. [Google Scholar] [PubMed]
- Mora, A.; Komander, D.; van Aalten, D.M.; Alessi, D.R. Pdk1, the master regulator of agc kinase signal transduction. Semin. Cell Dev. Biol. 2004, 161–170. [Google Scholar] [CrossRef]
- Mäemets-Allas, K.; Viil, J.; Jaks, V. A novel inhibitor of akt1–pdpk1 interaction efficiently suppresses the activity of akt pathway and restricts tumor growth in vivo. Mol. Cancer Ther. 2015, 14, 2486–2496. [Google Scholar] [CrossRef] [PubMed]
- Das, P.C.; Cao, Y.; Rose, R.L.; Cherrington, N.; Hodgson, E. Enzyme induction and cytotoxicity in human hepatocytes by chlorpyrifos and N,N-diethyl-m-toluamide (deet). Drug Metabol. Drug Interact. 2008, 23, 237–260. [Google Scholar] [CrossRef] [PubMed]
- LeCluyse, E.L.; Madan, A.; Hamilton, G.; Carroll, K.; DeHaan, R.; Parkinson, A. Expression and regulation of cytochrome p450 enzymes in primary cultures of human hepatocytes. J. Biochem. Mol. Toxicol. 2000, 14, 177–188. [Google Scholar] [CrossRef]
- Baselt, R.C. Disposition of Toxic Drugs and Chemicals in Man, 4th ed.; Preston Publications Inc.: Niles, IL, USA, 1994; ISBN 0962652318. [Google Scholar]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with tophat and cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J. Venny. An Interactive Tool for Comparing Lists with Venn Diagrams. 2007. Available online: http://www.citeulike.org/user/hroest/article/6994833 (accessed on 15 August 2017).
- Mudunuri, U.; Che, A.; Yi, M.; Stephens, R.M. Biodbnet: The biological database network. Bioinformatics 2009, 25, 555–556. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.N.; Goswami, R.; Pal, A. The insect repellents: A silent environmental chemical toxicant to the health. Environ. Toxicol. Pharmacol. 2017, 50, 91–102. [Google Scholar] [CrossRef] [PubMed]

| Differential Expression | Gene Symbol a | Gene Name b | Log2FC (DT) | Log2FC (Fip) | Log2FC (DT+Fip) |
|---|---|---|---|---|---|
| Up | CYP2B7P | Cytochrome P450 family 2 subfamily B member 7, pseudogene | +2.96 | +2.42 | +2.79 |
| HCP5 | HLA complex P5 | +0.78 | +0.93 | +1.38 | |
| Down | MALAT1 | Metastasis associated lung adenocarcinoma transcript 1 | −1.34 | −1.87 | −2.28 |
| NEAT1 | Nuclear paraspeckle assembly transcript 1 | −1.18 | −1.22 | −1.52 | |
| LINC01554 | Long intergenic non-protein coding RNA 1554 | −1.1 | −1.37 | −1.33 | |
| LINC01004 | Long intergenic non-protein coding RNA 1004 | −1.58 | −2.01 | −2.26 | |
| PSORS1C3 | Psoriasis susceptibility 1 candidate 3 | −1.58 | −0.85 | −1.39 | |
| AQP7P1 | Aquaporin 7 pseudogene 1 | −0.8 | −1.05 | −0.55 | |
| SCART1 | Scavenger receptor protein family member | −1.49 | −1.24 | −1.89 | |
| PDXDC2P | Pyridoxal dependent decarboxylase domain containing 2 | −0.78 | −1.14 | −1.41 | |
| LINC00893 | Long intergenic non-protein coding RNA 893 | −1.21 | −1.18 | −1.51 | |
| WASH5P | WAS protein family homolog 5 pseudogene | −0.79 | −0.76 | −1.06 | |
| PFN1P2 | Profilin 1 pseudogene 2 | −1.24 | −1.33 | −1.78 | |
| LINC00482 | Long intergenic non-protein coding RNA 482 | −1.01 | −0.91 | −1.06 | |
| ERVK13-1 | Endogenous retrovirus group K13 member 1 | −0.84 | −0.78 | −1.25 | |
| LOC100289230 | Uncharacterized LOC100289230 | −1.66 | −1.8 | −1.44 | |
| LOC728040 | HCG1813624 | −1.37 | −2.5 | −3.47 | |
| LOC100190986 | Uncharacterized LOC100190986 | −0.91 | −0.96 | −1.29 | |
| LINC01000 | Long intergenic non-protein coding RNA 1000 | −0.71 | −0.53 | −0.88 | |
| LOC100272217 | Uncharacterized LOC100272217 | −1.46 | −1.29 | −1.54 |
| lncRNA | Gene(s) within 1000 kb of lncRNA |
|---|---|
| CYP2B7P | CYP2A7 (−54710), CYP2B6 (−53837), CYP2A6 *, CYP2A13 * |
| HCP5 | MICB (−33621), MICA (+60915) |
| AQP7P1 | ANKRD20A1 (−646908) |
| MALAT1 | SCYL1 (−22962), FRMD8 (+115516), NEAT1 * |
| SCART1 | CYP2E1 (−59218), MTG1 (+67017) |
| PFN1P2 | PPIAL4B (−247525), NBPF9 (−199977) |
| PDXDC2P | PDPR (−92503), CLEC18A (+69943), NQO1 * |
| LINC01000 | CALU (−88173), METTL2B (+174390) |
| LOC100190986 | METTL9 (−166237), NPIPB3 (−13482) |
| PSORS1C3 | POU5F1 (−5124), HLA-C (+96269), HCP5 * |
| LINC01554 | GLRX (−33468), ELL2 (+105889) |
| LOC100272217 | FUBP3 (−1184) |
| LINC00893 | IDS (−28345), CXorf40A (−6965) |
| NEAT1 | SCYL1 (−100412), FRMD8 (+38066), MALAT1 * |
| WASH5P | OR4F17 (−41513) |
| LOC100289230 | CHD1 (−3535) |
| LINC00482 | SLC38A10 (−10731), TMEM105 (+24638) |
| LINC01004 | KMT2E (−27723), LHFPL3 (+657799) |
| ERVK13-1 | KCTD5 (−16561), PDPK1 (+127950), RPS2 * |
| LOC728040 | AFM (+36985), RASSF6 (+101963), CXCL8 * |
| Diff. Exp. | Gene Symbol a | Gene Name b | FC c (DT) | FC c (Fip) | FC c (DT+Fip) |
|---|---|---|---|---|---|
| Up | H19 | Long Intergenic Non-Protein Coding RNA 8 | - | +1.55 | +0.59 |
| Down | HULC | Highly Up-Regulated In Liver Cancer | - | −0.57 | - |
| MALAT1 | Metastasis Associated Lung Adenocarcinoma Transcript 1 | −1.34 | −1.87 | −2.28 | |
| NEAT1 | Nuclear Enriched Abundant Transcript 1 | −1.10 | −1.22 | −1.52 | |
| XIST | X (Inactive)-Specific Transcript | - | −0.60 | −0.93 | |
| TSIX | TSIX Transcript, XIST Antisense RNA | - | −0.59 | −0.97 | |
| MEG3 | Maternally Expressed 3 | - | −1.19 | −1.70 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitchell III, R.D.; Wallace, A.D.; Hodgson, E.; Roe, R.M. Differential Expression Profile of lncRNAs from Primary Human Hepatocytes Following DEET and Fipronil Exposure. Int. J. Mol. Sci. 2017, 18, 2104. https://doi.org/10.3390/ijms18102104
Mitchell III RD, Wallace AD, Hodgson E, Roe RM. Differential Expression Profile of lncRNAs from Primary Human Hepatocytes Following DEET and Fipronil Exposure. International Journal of Molecular Sciences. 2017; 18(10):2104. https://doi.org/10.3390/ijms18102104
Chicago/Turabian StyleMitchell III, Robert D., Andrew D. Wallace, Ernest Hodgson, and R. Michael Roe. 2017. "Differential Expression Profile of lncRNAs from Primary Human Hepatocytes Following DEET and Fipronil Exposure" International Journal of Molecular Sciences 18, no. 10: 2104. https://doi.org/10.3390/ijms18102104