Phytochemical Analysis of Agrimonia pilosa Ledeb, Its Antioxidant Activity and Aldose Reductase Inhibitory Potential
Abstract
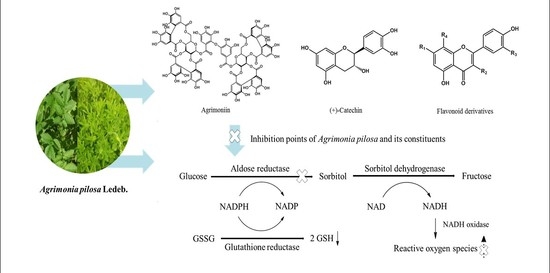
:1. Introduction
2. Results
2.1. Structural Determination of Isolated Compounds
2.2. Inhibitory Effect of Isolated Compounds on RLAR
2.3. DPPH and Off-Line DPPH HPLC Assay
2.4. Kinetic-Type RHAR Inhibition by the Active Compounds
2.5. Lens Culture and Intracellular Sorbitol Measurement
3. Discussion
4. Materials and Methods
4.1. General
4.2. Chemicals and Reagents
4.3. Plant Materials
4.4. Extraction, Fractionation, and Isolation
4.5. Preparation of Aldose Reductase
4.6. Determination of RLAR Inhibition In Vitro
4.7. HPLC Analysis
4.8. Evaluation of DPPH Radical Scavenging Capacity
4.9. OffLine DPPH HPLC Assay
4.10. Determination of Inhibition-Type of RHAR by Active Compound
4.11. Lens Culture and Intracellular Sorbitol Measurement
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AR | aldose reductase |
| NADPH | nicotinamide adenine dinucleotide phosphate |
| NADH | nicotinamide adenine dinucleotide |
| CT | catechin |
| AP | Agrimonia pilosa Ledeb |
| LT | luteolin |
| QC | quercetin |
| IQC | isoquercetin |
| HP | hyperin |
| AG | apigenin |
| VT | vitexin |
| KP | kaempferol |
| AS | astragalin |
| AZ | afzelin |
| RLAR | rat lens aldose reductase |
| APE | Agrimonia pilosa 50% methanol (MeOH) extract |
| RHAR | recombinant human aldose reductase |
| ICs | isolated compounds isolated from Agrimonia pilosa |
| KNCs | known compounds isolated from Agrimonia pilosa |
| TMG | tetramethylene glutaric acid |
| PAR | peak area reduction |
| CH2Cl2 | methylene chloride |
| EtOAc | ethyl acetate |
| n-BuOH | n-butanol |
| DPPH | 1,1-diphenyl-2-picrylhydrazyl |
References
- Fatmawati, S.; Kurashiki, K.; Takeno, S.; Kim, Y.U.; Shimizu, K.; Sato, M.; Imaizumi, K.; Takahashi, K.; Kamiya, S.; Kaneko, S.; et al. The inhibitory effect on aldose reductase by an extract of Ganoderma lucidum. Phytother. Res. 2009, 23, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Peyroux, J.; Sternberg, M. Advanced glycation endproducts (AGEs): Pharmacological inhibition in diabetes. Pathol. Biol. 2006, 54, 405–419. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi, M.; Ranjbar, A.; Shadnia, S.; Nikfar, S.; Rezaie, A. Pesticides and oxidative stress: A review. Med. Sci. Monit. 2004, 10, 141–147. [Google Scholar]
- Sundaram, R.K.; Bhaskar, A.; Vijayalingam, S.; Viswanathan, M.; Mohan, R.; Shanmugasundaram, K.R. Antioxidant status and lipid peroxidation in type II diabetes mellitus with and without complications. Clin. Sci. 1996, 90, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Matough, F.A.; Budin, S.B.; Hamid, Z.A.; Alwahaibi, N.; Mohamed, J. The role of oxidative stress and antioxidants in diabetic complications. Sultan Qaboos Univ. Med. J. 2012, 12, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Tana, J.; Wang, B.; Hea, R.; Liuc, Y.; Zhenga, C. Antioxidant activities of aqueous extract from Agrimonia pilosa Ledeb and its fractions. Chem. Biodivers. 2009, 6, 1716–1726. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Jiang, J.; Shim, D.W.; Kwon, S.C.; Kim, T.J.; Ye, S.K.; Kim, M.K.; Shin, Y.K.; Koppula, S.; Kang, T.B.; et al. Anti-inflammatory and anti-allergic effects of Agrimonia pilosa Ledeb extract on murine cell lines and OVA-induced airway inflammation. J. Ethnopharmacol. 2012, 140, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.K.; Park, M.S. Acetylcholinesterase inhibition by flavonoids from Agrimonia pilosa. Molecules 2007, 12, 2130–2139. [Google Scholar] [CrossRef] [PubMed]
- An, R.B.; Kim, H.C.; Jeong, G.S.; Oh, S.H.; Oh, H.; Kim, Y.C. Constituents of the aerial parts of Agrimonia pilosa. Nat. Prod. Sci. 2005, 11, 196–198. [Google Scholar]
- Jiang, Q.; Ma, J.; Wang, Y.; Ding, L.; Chen, L.; Qiu, F. Simultaneous determination of nine major constituents in Agrimonia pilosa Ledeb. by HPLC-DAD-ESI-MS/MS. Anal. Methods 2014, 6, 4373–4379. [Google Scholar] [CrossRef]
- Kato, H.; Li, W.; Koike, M.; Wang, Y.; Koike, K. Phenolic glycosides from Agrimonia pilosa. Phytochemistry 2010, 71, 1925–1929. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhu, L.; Tan, J.; Zhou, X.; Xiao, L.; Yang, X.; Wang, B. Glucosidase inhibitory activity and antioxidant activity of flavonoid compound and triterpenoid compound from Agrimonia pilosa Ledeb. BMC Complement. Altern. Med. 2014, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Veeresham, C.; Rama Rao, A.; Asres, K. Aldose reductase inhibitors of plant origin. Phytother. Res. 2013, 28, 317–333. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.A.; Isslam, M.D.N.; Kwon, Y.S.; Jin, S.E.; Son, Y.K.; Park, J.J.; Sohn, H.S.; Choi, J.S. Extraction and identification of three major aldose reductase inhibitors from Artemisia montana. Food Chem. Toxicol. 2011, 49, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Liu, Q.; Wang, M.; Jiang, M.; Jiang, S.; Zhang, L.; He, L.; Wang, J.; Chen, X. Target-guided separation of antioxidants from Semen cassia via off-line two-dimensional high-speed counter-current chromatography combined with complexation and extrusion elution mode. J. Chromatogr. B 2015, 1001, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Li, H.J.; Chen, J.; Guo, C.W.; Li, P. Rapid and simple method for screening of natural antioxidants from Chinese herb Flos Lonicerae japonicae by DPPH-HPLC-DAD-TOF/MS. J. Sep. Sci. 2008, 31, 3519–3526. [Google Scholar] [CrossRef] [PubMed]
- Olennikov, D.N.; Kashchenko, N.I.; Chirikova, N.K.; Kuzmina, S.S. Phenolic profile of Potentilla anserina L. (Rosaceae) herb of Siberian origin and development of a rapid method for simultaneous determination of major phenolics in P. anserina pharmaceutical products by microcolumn RP-HPLC-UV. Molecules 2015, 20, 224–248. [Google Scholar] [CrossRef] [PubMed]
- Fathiazad, F.; Delazar, A.; Amiri, R.; Sarker, S.D. Extraction of flavonoids and quantification of rutin from waste tobacco leaves. Iran. J. Pharm. Res. 2006, 3, 222–227. [Google Scholar]
- Lee, S.H.; Choi, M.J.; Choi, J.M.; Lee, S.; Kim, H.Y.; Cho, E.J. Flavonoids from Taraxacum coreanum protect from radical-induced oxidative damage. J. Med. Plants Res. 2012, 6, 5377–5384. [Google Scholar]
- Lu, Y.; Foo, L.Y. Flavonoid and phenolic glycosides from Salvia officinalis. Phytochemistry 2000, 55, 263–267. [Google Scholar] [CrossRef]
- Lu, Y.; Foo, L.Y. Identiücation and quantiücation of major polyphenols in apple pomace. Food Chem. 1997, 59, 187–194. [Google Scholar] [CrossRef]
- Pieroni, A.; Heimler, D.; Pieters, L.; van Poel, B.; Vlietnick, A.J. In vitro anti-complementary activity of flavonoids from olive (Olea europaea L.) leaves. Pharmazie 1996, 51, 765–768. [Google Scholar] [PubMed]
- Baris, O.; Karadayi, M.; Yanmis, D.; Guvenalp, Z.; Bal, T.; Gulluce, M. Isolation of 3 flavonoids from Mentha longifolia (L.) Hudson sucsp. longifolia and determination of their genotoxic potentials by using the E. coli WP2 test system. J. Food Sci. 2011, 76, 212–217. [Google Scholar]
- Matsuda, H.; Morikawa, T.; Toguchida, I.; Yoshikawa, M. Structural requirements of flavonoids and related compounds for aldose reductase inhibitory activity. Chem. Pharm. Bull. 2002, 50, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Naeem, S.; Hylands, P.; Barlow, D. Construction of an Indonesian herbal constituents database and its use in Random Forest modelling in a search for inhibitors of aldose reductase. Bioorg. Med. Chem. 2012, 20, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Li, H.M.; Hwang, S.H.; Kang, B.G.; Hong, J.S.; Lim, S.S. Inhibitory effects of Colocasia esculenta (L.) Schott constituents on aldose reductase. Molecules 2014, 19, 13212–13224. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, M.; Morikawa, T.; Murakami, T.; Toguchida, I.; Harima, S.; Matsuda, H. Medical flowers, I. Aldose reductase inhibitors and three new eudesmane-type sesquiterpenes, kikkanols A, B, and C, from the flowers of Chrysanthemum indicum L. Chem. Pharm. Bull. 1999, 47, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, H.; Morikawa, T.; Toguchida, I.; Harima, S.; Yoshikawa, M. Medicinal flowers. VI. Absolute stereostructures of two new flavanone glycosides and a phenylbutanoid glycoside from the Flowers of Chrysanthemum indicum L.: Their inhibitory activities for rat lens aldose reductase. Chem. Pharm. Bull. 2002, 50, 972–975. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, M.; Shimada, H.; Nishida, N.; Li, Y.; Toguchida, I.; Yamahara, J.; Matsuda, H. Antidiabetic principles of natural medicines. II. aldose reductase and α-glucosidase inhibitors from Brazilian natural medicine, the leaves of Myrcia multiflora DC. (Myrtacae): Structures of myrciacitrins I and II and myrciaphenones A and B. Chem. Pharm. Bull. 1998, 46, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Mok, S.Y.; Lee, S.H. Identification of flavonoids and flavonoid rhamnosides from Rhododendron mucronulatum for albiflorum and their inhibitory activities against aldose reductase. Food Chem. 2013, 136, 969–974. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.H.; Song, D.G.; Lee, J.Y.; Pan, C.H.; Um, B.H.; Jung, S.H. Flavonoids from the leaves of Thuja orientalis inhibit the aldose reductase and the formation of advanced glycation endproducts. J. Korean Soc. Appl. Biol. Chem. 2009, 52, 448–455. [Google Scholar] [CrossRef]
- Ha, T.J.; Lee, J.H.; Lee, M.H.; Lee, B.W.; Kwon, H.S.; Park, C.H.; Shim, K.B.; Kim, H.T.; Baek, I.Y.; Jang, D.S. Isolation and identification of phenolic compounds from the seeds of Perilla frutescens (L.) and their inhibitory activities against α-glucosidase and aldose reductase. Food Chem. 2012, 135, 1397–1403. [Google Scholar] [CrossRef] [PubMed]
- Chethan, S.; Dharmesh, S.M.; Malleshi, N.G. Inhibition of aldose reductase from cataracted eye lenses by finger millet (Eleusine coracana) polyphenols. Bioorg. Med. Chem. 2008, 16, 10085–10090. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.P.; Shi, S.Y.; Xiong, X.; Chen, X.Q.; Peng, M.J. Comparative evaluation of three methods based on high-performance liquid chromatography analysis combined with a 2,2′-diphenyl-1-picrylhydrazyl assay for the rapid screening of antioxidants from Pueraria lobate flowers. Anal. Bioanal. Chem. 2012, 402, 2965–2976. [Google Scholar] [CrossRef] [PubMed]
- Dia, X.; Huang, Q.H.; Zhou, B.; Gong, Z.; Liu, Z.; Shi, S. Preparative isolation and purification of seven main antioxidants from Eucommia ulmoides Oliv. (Du-zhong) leaves using HSCCC guided by DPPH-HPLC experiment. Food Chem. 2013, 139, 563–570. [Google Scholar]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.N.; Lee, M.Y.; Kim, J.K.; Suh, H.W.; Lim, S.S. Aldose reductase inhibitory compounds from Xanthium strumarium. Arch. Pharm. Res. 2013, 36, 1090–1095. [Google Scholar] [CrossRef] [PubMed]
- Peak, J.H.; Lim, S.S. Preparative isolation of aldose reductase inhibitory compounds from Nardostachys chinensis by elution-extrusion counter-current chromatography. Arch. Pharm. Res. 2014, 37, 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.W.; Kim, S.C.; Hwang, S.S.; Choi, B.K.; Ahn, H.J.; Lee, M.Y.; Park, S.H.; Kim, S.K. Antioxidant activity and free radical scavenging capacity between Korean medicinal plants and flavonoids by assay-guided comparison. Plant Sci. 2002, 163, 1161–1168. [Google Scholar] [CrossRef]
- Shi, S.Y.; Ma, Y.J.; Zhang, Y.P.; Liu, L.L.; Liu, Q.; Peng, M.J.; Xiong, X. Systematic separation and purification of 18 antioxidants from Pueraria lobata flower using HSCCC target-guided by DPPH-HPLC experiment. Sep. Purif. Technol. 2012, 89, 225–233. [Google Scholar] [CrossRef]
- Kim, T.H.; Kim, J.K.; Kang, Y.H.; Lee, J.Y.; Kang, I.J.; Lim, S.S. Aldose reductase inhibitory activity of compounds from Zea mays L. Biomed. Res. Int. 2013, 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Kim, S.H.; Jung, S.H.; Kim, J.K.; Pan, C.H.; Lim, S.S. Aldose reductase inhibitory compounds from Glycyrrhiza uralensis. Biol. Pharm. Bull. 2010, 33, 917–921. [Google Scholar] [CrossRef] [PubMed]





| Extract and Fractions | Inhibition (%) | ||
|---|---|---|---|
| RLAR | DPPH | ||
| Methylene chloride extract | 6.8 ± 0.20 | 4.19 ± 0.14 | |
| 50% MeOH ext. | Crude extract | 51.4 ± 0.10 | 53.4 ± 0.14 |
| EtOAc fraction | 84.4 ± 0.27 | 62.3 ± 0.04 | |
| n-BuOH fraction | 92.4 ± 0.14 | 61.0 ± 0.42 | |
| Water fraction | 37.9 ± 0.47 | 33.0 ± 0.10 | |
| RLAR | TMG | 99.7 ± 0.11 | - |
| DPPH | Ascorbic acid | - | 81.0 ± 0.01 |
| Entry | Compounds | DPPH | RLAR | |||
|---|---|---|---|---|---|---|
| Experiments | References | |||||
| IC50 (μM) | Inhibition (%) | IC50 (μM) | IC50 (μM) | |||
| IC a) | Agrimoniin (AM) | 13.0 ± 0.06 | 35.0 ± 0.41 | 1.6 ± 0.12 | - | |
| Rutin (RT) | 66.8 ± 0.34 | 31.7 ± 0.65 | 9.5 ± 0.75 | 9.0 [24] b) | ||
| Luteolin-7-O-glucoside (LGC) | 71.5 ± 0.29 | 46.9 ± 0.95 | 8.1 ± 0.72 | 7.5 [26] | ||
| Luteolin-7-O-glucuronide (LGN) | 80.6 ± 0.38 | 83.3 ± 0.88 | 0.7 ±0.54 | 3.1 [27] | ||
| Apigenin-7-O-glucoside (AGC) | >250 | 40.2 ± 0.56 | 4.3 ± 0.14 | 23.0 [28] | ||
| Quercitrin (QU) | 77.9 ± 0.27 | 97.4 ± 1.38 | 0.2 ±0.02 | 0.2 [29] | ||
| Apigenin-7-O-glucuronide (AGN) | >250 | <0 | >30 | - | ||
| KNC b) | Catechin (CT) | 106.7 ± 0.43 | 7.2 ± 1.02 | >30 | >30 [24] | |
| Kaempferol (KP) | 91.6 ± 0.68 | 11.8 ± 0.81 | 15.2 ± 1.32 | 10 [24] | ||
| Quercetin (QC) | 70.4 ± 0.15 | 74.1 ± 0.85 | 3.2 ± 0.13 | 2.2 [24] | ||
| Isoquercitrin (IQC) | 65.9 ± 0.46 | 41.0 ± 1.07 | 5.1 ± 0.88 | 4.5 [24] | ||
| Hyperin (HP) | 73.3 ± 0.23 | 90.8 ± 0.96 | 4.1 ± 0.32 | 3.0 [24] | ||
| Apigenin (AG) | 156.3 ± 1.21 | 81.8 ± 1.20 | 3.2 ± 0.21 | 2.2 [24] | ||
| Vitexin (VT) | >250 | 12.2 ± 0.95 | >30 | >30 [25] | ||
| Astragalin (AS) | >250 | 53.3 ± 1.14 | 5.1 ± 0.89 | >30 [25] | ||
| Luteolin (LT) | 88.2 ± 0.52 | 80.2 ± 0.90 | 0.6 ± 0.03 | 0.5 [27] | ||
| Afzelin (AZ) | >250 | 86.2 ± 0.38 | 1.0 ± 0.27 | 0.3 [30] | ||
| Positive control | DPPH | l-Ascorbic acid | 147.3 ± 0.43 | - | - | - |
| RLAR | TMG | - | 119.7 ± 0.22 | 0.5 ± 0.05 | 1.0 [30] | |
| Compounds | Sorbitol Content (mg)/Lens Wet Weight (g) a) | Inhibition (%) | Inhibition Types (References) |
|---|---|---|---|
| Sorbitol free | No detection | - | - |
| Control | 1.47 ± 0.04 | ||
| Quercetin a) | 0.21 ± 0.02 | 85.7 ± 8.32 | Noncompetitive [31] |
| Agrimoniin (AM) | 0.77 ± 0.02 | 47.6 ± 1.34 | Noncompetitive |
| Luteolin-7-O-glucuronide (LGN) | 0.12 ± 0.01 | 91.8 ± 9.01 | Noncompetitive |
| Quercitrin (QU) | 0.34 ± 0.02 | 76.9 ± 5.32 | Uncompetitive [29] |
| Luteolin (LT) | 0.12 ± 0.01 | 91.8 ± 7.91 | Mixed type [30] |
| Afzelin (AZ) | 0.10 ± 0.01 | 93.2 ± 8.67 | Noncompetitive |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.B.; Hwang, S.H.; Suh, H.-W.; Lim, S.S. Phytochemical Analysis of Agrimonia pilosa Ledeb, Its Antioxidant Activity and Aldose Reductase Inhibitory Potential. Int. J. Mol. Sci. 2017, 18, 379. https://doi.org/10.3390/ijms18020379
Kim SB, Hwang SH, Suh H-W, Lim SS. Phytochemical Analysis of Agrimonia pilosa Ledeb, Its Antioxidant Activity and Aldose Reductase Inhibitory Potential. International Journal of Molecular Sciences. 2017; 18(2):379. https://doi.org/10.3390/ijms18020379
Chicago/Turabian StyleKim, Set Byeol, Seung Hwan Hwang, Hong-Won Suh, and Soon Sung Lim. 2017. "Phytochemical Analysis of Agrimonia pilosa Ledeb, Its Antioxidant Activity and Aldose Reductase Inhibitory Potential" International Journal of Molecular Sciences 18, no. 2: 379. https://doi.org/10.3390/ijms18020379