Expression of Iron-Related Proteins Differentiate Non-Cancerous and Cancerous Breast Tumors
Abstract
:1. Introduction
2. Results
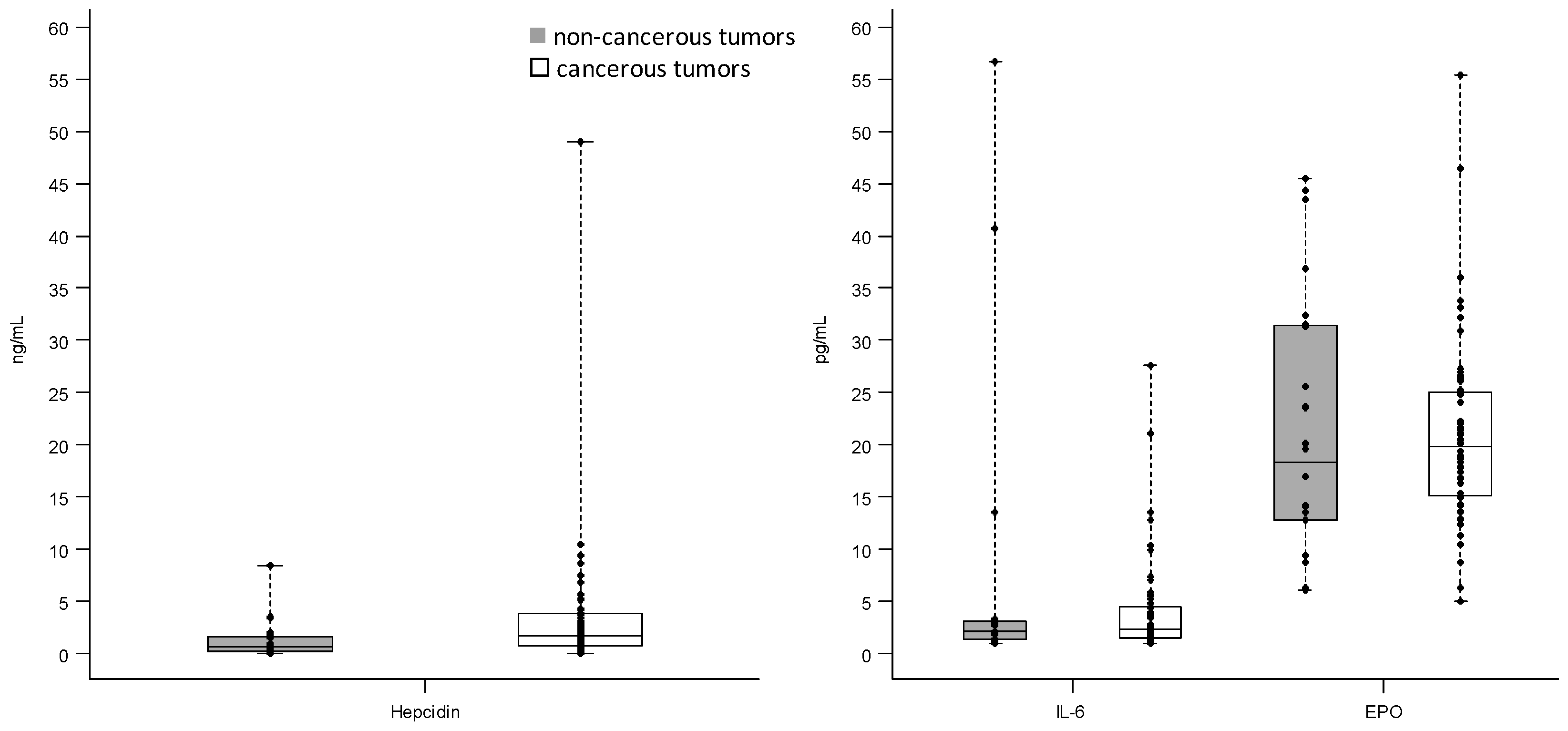
2.1. Hepcidin, Interleukin-6 and Erythropoietin Levels in Breast Cancerous and Non-Cancerous Tumor Tissues
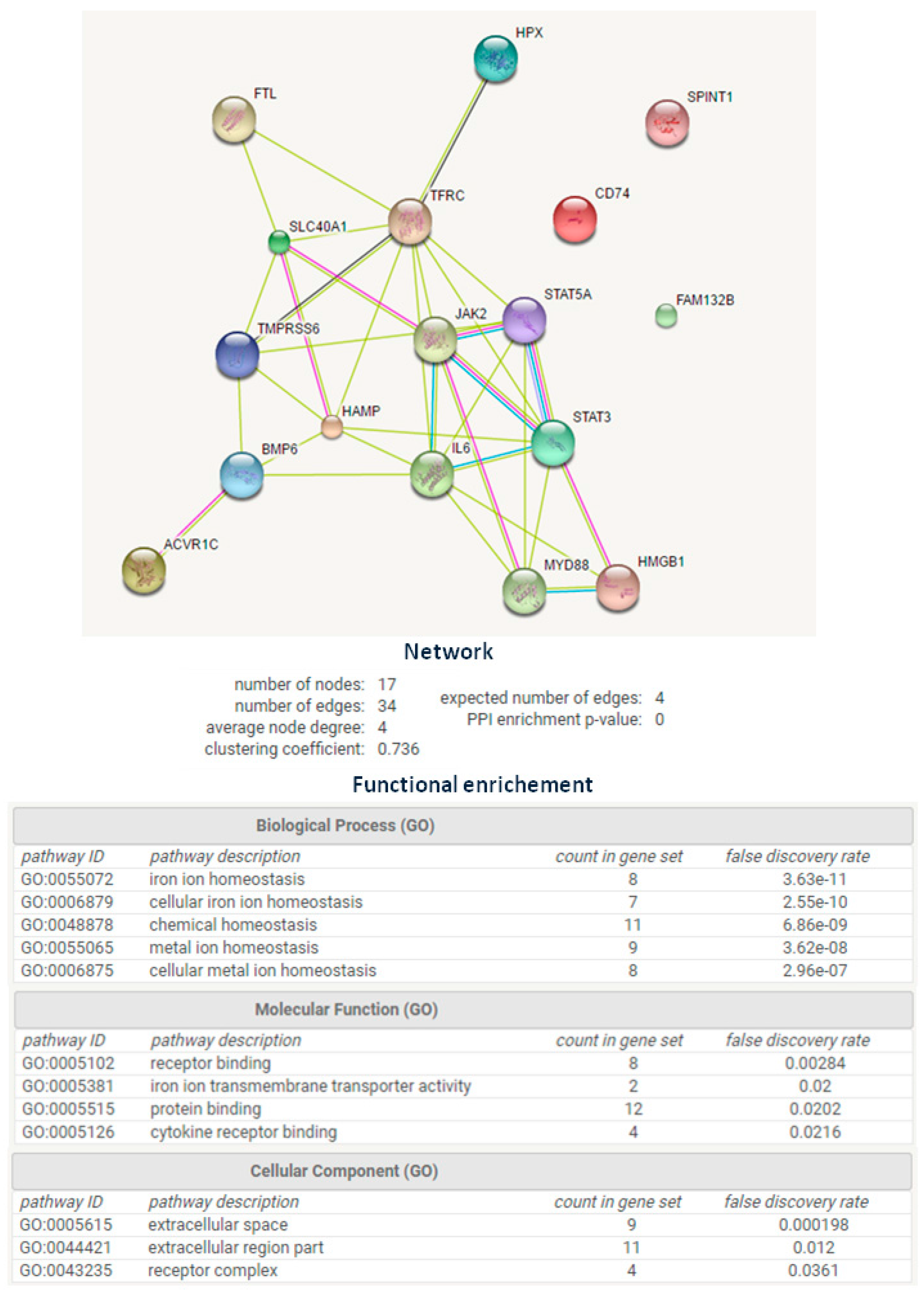
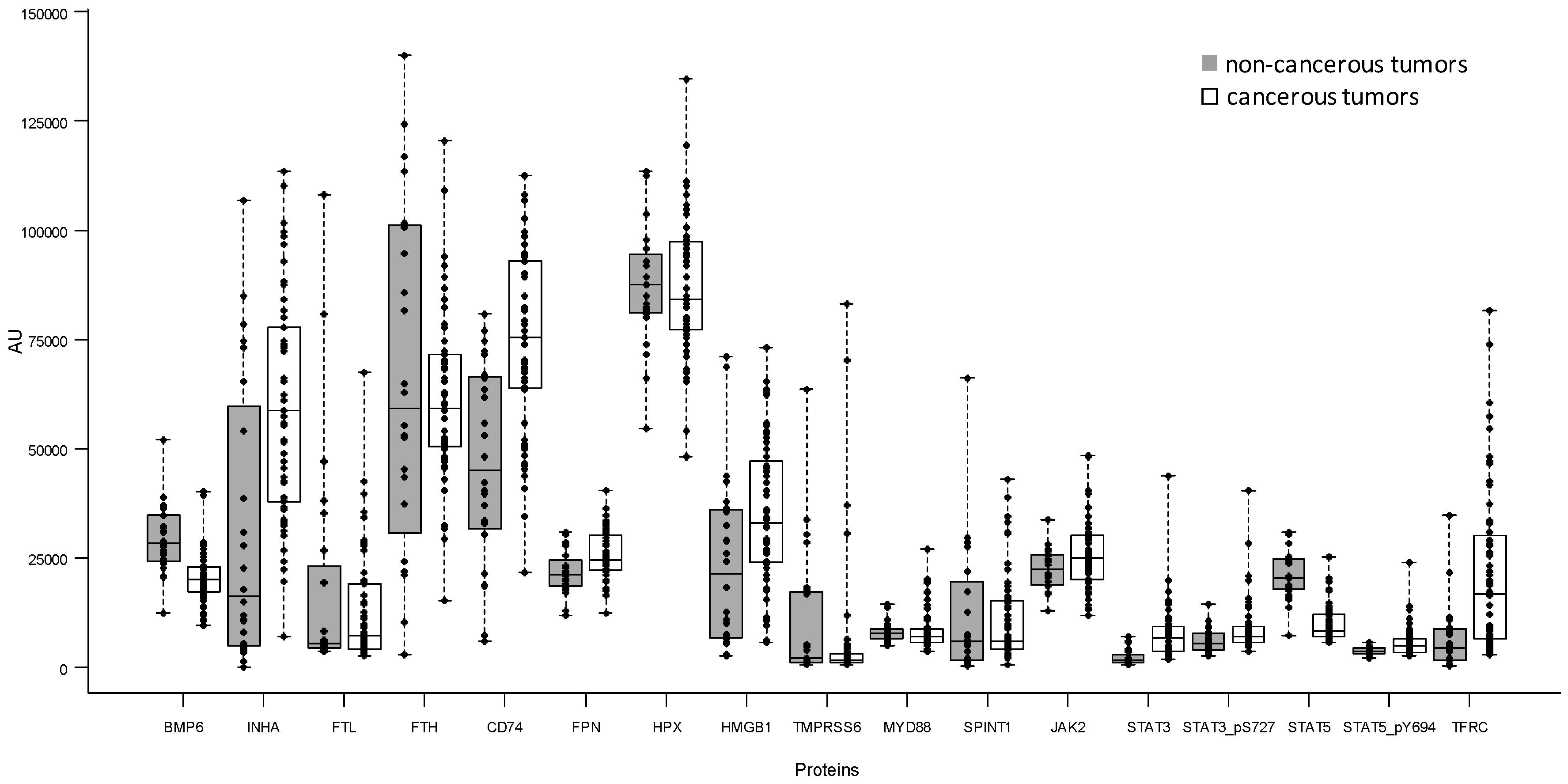
2.2. Comparison of 17 Iron-Related Proteins Measured by Reverse-Phase Protein Array between Non-Cancerous and Cancerous Tumors
2.2.1. Univariate Analysis
2.2.2. Multivariate Analysis
2.2.3. Selection of Relevant Proteins for Discriminate Between Non-Cancerous and Cancerous Tumors
2.2.4. Exploratory Evaluation of the Expression of TFRC, FPN, FTL, and FTH with the Clinicopathological Data
3. Discussion
4. Materials and Methods
4.1. Samples Characteristics
4.2. Enzyme-Linked Immunosorbent Assay
4.3. Reverse-Phase Protein Array
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kwok, J.C.; Richardson, D.R. The iron metabolism of neoplastic cells: Alterations that facilitate proliferation? Crit. Rev. Oncol. Hematol. 2002, 42, 65–78. [Google Scholar] [CrossRef]
- Marques, O.; Porto, G.; Rema, A.; Faria, F.; Cruz Paula, A.; Gomez-Lazaro, M.; Silva, P.; Martins da Silva, B.; Lopes, C. Local iron homeostasis in the breast ductal carcinoma microenvironment. BMC Cancer 2016, 16, 187. [Google Scholar] [CrossRef] [PubMed]
- Jezequel, P.; Campion, L.; Spyratos, F.; Loussouarn, D.; Campone, M.; Guerin-Charbonnel, C.; Joalland, M.P.; Andre, J.; Descotes, F.; Grenot, C.; et al. Validation of tumor-associated macrophage ferritin light chain as a prognostic biomarker in node-negative breast cancer tumors: A multicentric 2004 National PHRC Study. Int. J. Cancer 2012, 131, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Alkhateeb, A.A.; Connor, J.R. The significance of ferritin in cancer: Anti-oxidation, inflammation and tumorigenesis. Biochim. Biophys. Acta 2013, 1836, 245–254. [Google Scholar] [CrossRef]
- Mertens, C.; Akam, E.A.; Rehwald, C.; Brune, B.; Tomat, E.; Jung, M. Intracellular iron chelation modulates the macrophage iron phenotype with consequences on tumor progression. PLoS ONE 2016, 11, e0166164. [Google Scholar] [CrossRef] [PubMed]
- Orlandi, R.; de Bortoli, M.; Ciniselli, C.M.; Vaghi, E.; Caccia, D.; Garrisi, V.; Pizzamiglio, S.; Veneroni, S.; Bonini, C.; Agresti, R.; et al. Hepcidin and ferritin blood level as noninvasive tools for predicting breast cancer. Ann. Oncol. 2014, 25, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Marcus, D.M.; Zinberg, N. Measurement of serum ferritin by radioimmunoassay: Results in normal individuals and patients with breast cancer. J. Natl. Cancer Inst. 1975, 55, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, A.; Jones, B.; Ricketts, C.; Bulbrook, R.D.; Wang, D.Y. Serum ferritin concentration in early breast cancer. Br. J. Cancer 1976, 34, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T.; Nemeth, E. Iron Homeostasis in host defence and inflammation. Nat. Rev. Immunol. 2015, 15, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Kali, A.; Charles, M.V.; Seetharam, R.S. Hepcidin—A novel biomarker with changing trends. Pharmacogn. Rev. 2015, 9, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Kautz, L.; Jung, G.; Valore, E.V.; Rivella, S.; Nemeth, E.; Ganz, T. Identification of erythroferrone as an erythroid regulator of iron metabolism. Nat. Genet. 2014, 46, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Ciniselli, C.M.; de Bortoli, M.; Taverna, E.; Varinelli, L.; Pizzamiglio, S.; Veneroni, S.; Bonini, C.; Orlandi, R.; Verderio, P.; Bongarzone, I. Plasma hepcidin in early-stage breast cancer patients: No relationship with interleukin-6, erythropoietin and erythroferrone. Expert Rev. Proteom. 2015, 12, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Lui, G.Y.; Kovacevic, Z.; Richardson, V.; Merlot, A.M.; Kalinowski, D.S.; Richardson, D.R. Targeting cancer by binding iron: Dissecting cellular signaling pathways. Oncotarget 2015, 6, 18748–18779. [Google Scholar] [CrossRef] [PubMed]
- Torti, S.V.; Torti, F.M. Iron and cancer: More ore to be mined. Nat. Rev. Cancer 2013, 13, 342–355. [Google Scholar] [CrossRef] [PubMed]
- Marziali, G.; Signore, M.; Buccarelli, M.; Grande, S.; Palma, A.; Biffoni, M.; Rosi, A.; d’Alessandris, Q.G.; Martini, M.; Larocca, L.M.; et al. Metabolic/proteomic signature defines two glioblastoma subtypes with different clinical outcome. Sci. Rep. 2016, 6, 21557. [Google Scholar] [CrossRef] [PubMed]
- STRING Software. Available online: http://string-db.org/ (accessed on 18 November 2016).
- Maurer, E.; Gutschow, M.; Stirnberg, M. Hepatocyte growth factor activator inhibitor type 2 (HAI-2) modulates hepcidin expression by inhibiting the cell surface protease matriptase-2. Biochem. J. 2013, 450, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Peterson, M.; Miller, J. Antigen presentation enhanced by the alternatively spliced invariant chain gene product P41. Nature 1992, 357, 596–598. [Google Scholar] [CrossRef] [PubMed]
- Lamy, P.J.; Durigova, A.; Jacot, W. Iron Homeostasis and anemia markers in early breast cancer. Clin. Chim. Acta 2014, 434, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G. Targeting cancer metabolism: A therapeutic window opens. Nat. Rev. Drug Discov. 2011, 10, 671–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simcox, J.A.; McClain, D.A. Iron and diabetes risk. Cell. Metab. 2013, 17, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Hatcher, H.C.; Singh, R.N.; Torti, F.M.; Torti, S.V. Synthetic and natural iron chelators: Therapeutic potential and clinical use. Future Med. Chem. 2009, 1, 1643–1670. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T.; Nemeth, E. Iron sequestration and anemia of inflammation. Semin. Hematol. 2009, 46, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Cremona, M.; Espina, V.; Caccia, D.; Veneroni, S.; Colecchia, M.; Pierobon, M.; Deng, J.; Mueller, C.; Procopio, G.; Lanzi, C.; et al. Stratification of clear cell renal cell carcinoma by signaling pathway analysis. Expert Rev. Proteom. 2014, 11, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Habashy, H.O.; Powe, D.G.; Staka, C.M.; Rakha, E.A.; Ball, G.; Green, A.R.; Aleskandarany, M.; Paish, E.C.; Douglas Macmillan, R.; Nicholson, R.I.; et al. Transferrin receptor (CD71) is a marker of poor prognosis in breast cancer and can predict response to tamoxifen. Breast Cancer Res. Treat. 2010, 119, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Ruan, K.; Song, G.; Ouyang, G. Role of hypoxia in the hallmarks of human cancer. J. Cell. Biochem. 2009, 107, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, F. Iron Homeostasis and tumorigenesis: Molecular mechanisms and therapeutic opportunities. Protein Cell 2015, 6, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Peck, A.R.; Witkiewicz, A.K.; Liu, C.; Stringer, G.A.; Klimowicz, A.C.; Pequignot, E.; Freydin, B.; Tran, T.H.; Yang, N.; Rosenberg, A.L.; et al. Loss of nuclear localized and tyrosine phosphorylated STAT5 in breast cancer predicts poor clinical outcome and increased risk of antiestrogen therapy failure. J. Clin. Oncol. 2011, 29, 2448–2458. [Google Scholar] [CrossRef] [PubMed]
- Khandekar, M.J.; Cohen, P.; Spiegelman, B.M. Molecular mechanisms of cancer development in obesity. Nat. Rev. Cancer 2011, 11, 886–895. [Google Scholar] [CrossRef] [PubMed]
- Andriopoulos, B., Jr.; Corradini, E.; Xia, Y.; Faasse, S.A.; Chen, S.; Grgurevic, L.; Knutson, M.D.; Pietrangelo, A.; Vukicevic, S.; Lin, H.Y.; et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat. Genet. 2009, 41, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Shpyleva, S.I.; Tryndyak, V.P.; Kovalchuk, O.; Starlard-Davenport, A.; Chekhun, V.F.; Beland, F.A.; Pogribny, I.P. Role of ferritin alterations in human breast cancer cells. Breast Cancer Res. Treat. 2011, 126, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Barekati, Z.; Radpour, R.; Lu, Q.; Bitzer, J.; Zheng, H.; Toniolo, P.; Lenner, P.; Zhong, X.Y. Methylation signature of lymph node metastases in breast cancer patients. BMC Cancer 2012, 12, 244. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Deng, Z.; Hatcher, H.; Miller, L.D.; Di, X.; Tesfay, L.; Sui, G.; d’Agostino, R.B., Jr.; Torti, F.M.; Torti, S.V. IRP2 regulates breast tumor growth. Cancer Res. 2014, 74, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Gabrielsen, J.S.; Gao, Y.; Simcox, J.A.; Huang, J.; Thorup, D.; Jones, D.; Cooksey, R.C.; Gabrielsen, D.; Adams, T.D.; Hunt, S.C.; et al. Adipocyte iron regulates adiponectin and insulin sensitivity. J. Clin. Investig. 2012, 122, 3529–3540. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, J.K.; Chakraborti, S. Nonparametric Statistical Inference, 4th ed.; Marcel Dekker, INC: New York, NY, USA; Basel, Switzerland, 2003; pp. 231–280. [Google Scholar]
- Hosmer, D.W.; Lemeshow, S. Applied Logistic Regression, 3rd ed.; John Wiley & Sons: New York, NY, USA, 1989; pp. 47–77. [Google Scholar]
- Durrleman, S.; Simon, R. Flexible regression models with cubic splines. Stat Med. 1989, 8, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Hardle, W.K.; Leopold, S. Applied Multivariate Statistical Analysis, 2nd ed.; Springer-Verlag: Heidelberg, Germany, 2003; pp. 289–300. [Google Scholar]

| Proteins | Tumor Lesion | N | Minimum | Median | Maximum | IQR 1 |
|---|---|---|---|---|---|---|
| Hepcidin (ng/mL) | non-cancerous | 19 | 0.002 | 0.60 | 8.40 | 1.58 |
| cancerous | 52 | 0.03 | 1.75 | 48.97 | 3.00 | |
| IL-6 (pg/mL) | non-cancerous | 19 | 1.01 | 2.15 | 56.70 | 1.84 |
| cancerous | 50 | 1.01 | 2.32 | 27.64 | 2.98 | |
| EPO (pg/mL) | non-cancerous | 24 | 6.04 | 18.29 | 45.48 | 18.65 |
| cancerous | 56 | 5.01 | 19.78 | 55.35 | 9.92 |
| Proteins | Tumor Lesion | N | Minimum | Median | Maximum | IQR 1 |
|---|---|---|---|---|---|---|
| BMP6 | non-cancerous | 24 | 12,259.0 | 28,427.5 | 52,052 | 10,662.5 |
| cancerous | 56 | 9672.0 | 20,090.5 | 40,135 | 5800.0 | |
| INHA | non-cancerous | 24 | 20.9 | 16,370.5 | 106,938 | 54,852.5 |
| cancerous | 56 | 7016.0 | 58,689.0 | 113,550 | 39,665.5 | |
| FTL | non-cancerous | 24 | 3558.0 | 5547.0 | 108,012 | 18,734.5 |
| cancerous | 56 | 2492.0 | 7292.0 | 67,508 | 15,029.0 | |
| FTH | non-cancerous | 24 | 2922.0 | 59,107.5 | 140,084 | 70,455.0 |
| cancerous | 56 | 15,093.0 | 59,281.5 | 120,572 | 21,168.0 | |
| CD74 | non-cancerous | 24 | 5843.0 | 45,121.5 | 80,822 | 34,907.0 |
| cancerous | 56 | 21,547.0 | 75,358.0 | 112,420 | 29,071.0 | |
| FPN | non-cancerous | 24 | 11,980.0 | 21,205.0 | 30,946 | 5768.0 |
| cancerous | 56 | 12,432.0 | 24,588.0 | 40,538 | 7937.5 | |
| HPX | non-cancerous | 24 | 54,721.0 | 87,553.0 | 113,550 | 13,154.5 |
| cancerous | 56 | 48,050.0 | 84,120.0 | 134,592 | 19,981.0 | |
| HMGB1 | non-cancerous | 24 | 2592.0 | 21,306.5 | 70,969 | 29,248.0 |
| cancerous | 56 | 5693.0 | 33,034.5 | 73,130 | 23,245.5 | |
| TMPRSS6 | non-cancerous | 24 | 561.0 | 2135.0 | 63,577 | 16,252.0 |
| cancerous | 56 | 670.0 | 1686.5 | 83,283 | 2000.5 | |
| MYD88 | non-cancerous | 24 | 4949.0 | 7820.5 | 14,343 | 2280.5 |
| cancerous | 56 | 3569.0 | 6995.0 | 27,174 | 2887.0 | |
| JAK2 | non-cancerous | 24 | 12,965.0 | 22,371.5 | 33,860 | 6968.0 |
| cancerous | 56 | 11,790.0 | 25,085.5 | 48,533 | 9799.0 | |
| SPINT1 | non-cancerous | 24 | 180.0 | 5993.0 | 66,171 | 17,960.0 |
| cancerous | 56 | 632.0 | 5913.5 | 43,045 | 11,015.0 | |
| STAT3 | non-cancerous | 24 | 605.0 | 1685.5 | 7087 | 1643.5 |
| cancerous | 56 | 1779.0 | 6741.5 | 43,914 | 5621.5 | |
| STAT3_pS727 | non-cancerous | 24 | 2492.0 | 5564.0 | 14,400 | 3766.5 |
| cancerous | 56 | 3548.0 | 7009.0 | 40,538 | 3507.5 | |
| STAT5 | non-cancerous | 24 | 7208.0 | 20,465.5 | 30,946 | 7034.5 |
| cancerous | 56 | 5745.0 | 8304.0 | 25,336 | 5038.0 | |
| STAT5_pY694 | non-cancerous | 24 | 2055.0 | 3554.5 | 5625 | 1425.5 |
| cancerous | 56 | 2660.0 | 4806.0 | 23,861 | 3023.5 | |
| TFRC | non-cancerous | 24 | 317.0 | 4293.5 | 34,892 | 7021.5 |
| cancerous | 56 | 2954.0 | 16,863.5 | 81,634 | 23,502.0 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pizzamiglio, S.; De Bortoli, M.; Taverna, E.; Signore, M.; Veneroni, S.; Cho, W.C.-s.; Orlandi, R.; Verderio, P.; Bongarzone, I. Expression of Iron-Related Proteins Differentiate Non-Cancerous and Cancerous Breast Tumors. Int. J. Mol. Sci. 2017, 18, 410. https://doi.org/10.3390/ijms18020410
Pizzamiglio S, De Bortoli M, Taverna E, Signore M, Veneroni S, Cho WC-s, Orlandi R, Verderio P, Bongarzone I. Expression of Iron-Related Proteins Differentiate Non-Cancerous and Cancerous Breast Tumors. International Journal of Molecular Sciences. 2017; 18(2):410. https://doi.org/10.3390/ijms18020410
Chicago/Turabian StylePizzamiglio, Sara, Maida De Bortoli, Elena Taverna, Michele Signore, Silvia Veneroni, William Chi-shing Cho, Rosaria Orlandi, Paolo Verderio, and Italia Bongarzone. 2017. "Expression of Iron-Related Proteins Differentiate Non-Cancerous and Cancerous Breast Tumors" International Journal of Molecular Sciences 18, no. 2: 410. https://doi.org/10.3390/ijms18020410






