Determination of Genes Related to Uveitis by Utilization of the Random Walk with Restart Algorithm on a Protein–Protein Interaction Network
Abstract
:1. Introduction
2. Results and Discussion
2.1. Results of Testing Random Walk with Restart (RWR)-Based Method
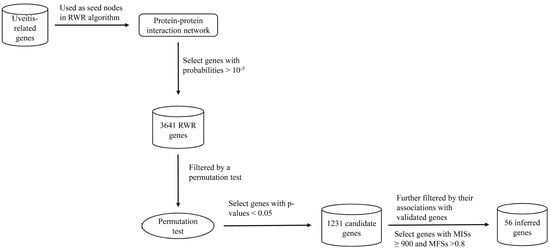
2.2. RWR Genes
2.3. Candidate Genes
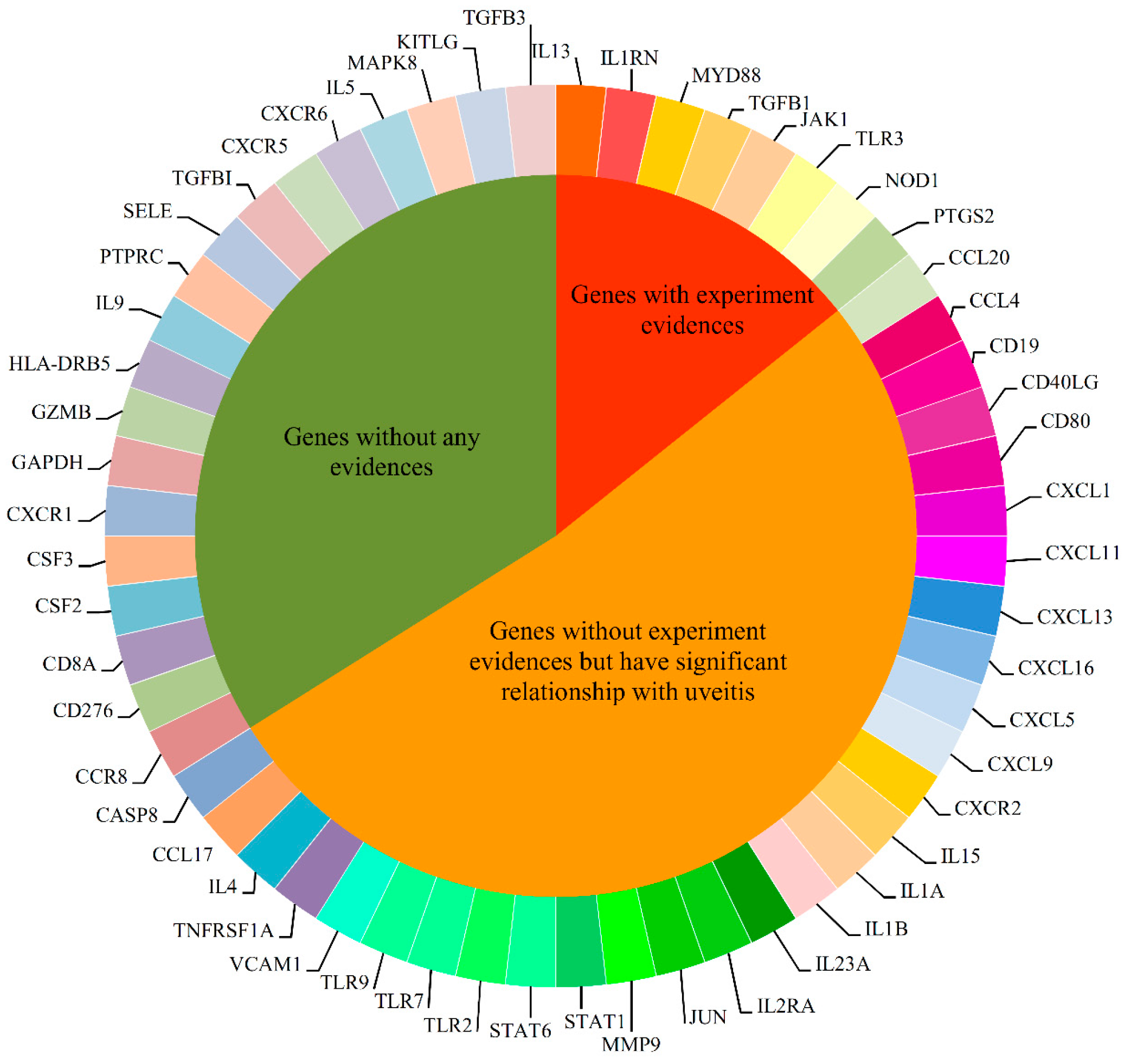
2.4. Analysis of Novel Genes
2.4.1. Immune System Regulation Genes
2.4.2. Transcription Associated Genes
2.4.3. Cell Adhesion and Signal Transduction Related Genes
2.5. Comparison of Other Methods
3. Materials and Methods
3.1. Materials
3.2. Protein-protein Interaction (PPI) Network
3.3. RWR-Based Method
3.3.1. Searching Possible Genes Using the RWR Algorithm
3.3.2. Excluding False Discoveries Using the Permutation Test
3.3.3. Selection of Core Genes by Associations with Validated Genes
3.4. Methods for Testing RWR-Based Method
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Miserocchi, E.; Foqliato, G.; Modorati, G.; Bandello, F. Review on the worldwide epidemiology of uveitis. Eur. J. Ophthalmol. 2013, 23, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Fraga, N.A.; Oliveira, M.F.; Follador, I.; Rocha, B.O.; Rêgo, V.R. Psoriasis and uveitis: A literature review. An. Bras. Dermatol. 2012, 87, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, P. Review: Uveitis and immunosuppressive drugs. J. Ocul. Pharmacol. Ther. 2001, 17, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Inaba, Y.; Mochizuki, M.; Nakajima, A.; Urayama, A. A nation-wide survey on the occurrence of Vogt-Koyanagi-Harada disease in Japan. Jpn. J. Ophthalmol. 1994, 98, 389–392. [Google Scholar]
- Moorthy, R.S.; Inomata, H.; Rao, N.A. Vogt-Koyanagi-Harada syndrome. Surv. Ophthalmol. 1995, 39, 265–292. [Google Scholar] [CrossRef]
- Evereklioglu, C. Current concepts in the etiology and treatment of Behcet disease. Surv. Ophthalmol. 2005, 50, 297–350. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.H.; Wakefield, D. Uveitis: A global perspective. Ocul. Immunol. Inflamm. 2002, 10, 263–279. [Google Scholar] [CrossRef] [PubMed]
- Priem, H.A.; Kijlstra, A.; Noens, L.; Baarsma, G.S.; de Laey, J.J.; Oosterhuis, J.A. HLA typing in birdshot chorioretinopathy. Am. J. Ophthalmol. 1988, 105, 182–185. [Google Scholar] [CrossRef]
- Chaput, F.; Amer, R.; Baglivo, E.; Touitou, V.; Kozyreff, A.; Bron, D.; Bodaghi, B.; LeHoang, P.; Bergstrom, C.; Grossniklaus, H.E.; et al. Intraocular T-cell Lymphoma: Clinical Presentation, Diagnosis, Treatment, and Outcome. Ocul. Immunol. Inflamm. 2016, 22, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kitazawa, K.; Nagata, K.; Yamanaka, Y.; Kuwahara, Y.; Lehara, T.; Kinoshita, S.; Sotozono, C. Diffuse Anterior Retinoblastoma with Sarcoidosis-Like Nodule. Case Rep. Ophthalmol. 2015, 6, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Catala-Mora, J.; Parareda-Salles, A.; Vicuña-Muñoz, C.G.; Medina-Zurinaga, M.; Prat-Bartomeu, J. [Uveitis masquerade syndrome as a presenting form of diffuse retinoblastoma]. Arch. Soc. Esp. Oftalmol. 2009, 84, 477–480. [Google Scholar] [PubMed]
- All-Ericsson, C.; Economou, M.A.; Landau, I.; Seregard, S.; Träisk, F. Uveitis masquerade syndromes: Diffuse retinoblastoma in an older child. Acta Ophthalmol. Scand. 2007, 85, 569–570. [Google Scholar] [CrossRef] [PubMed]
- Jovanovic, S.; Jovanović, Z.; Paović, J.; Teperković, V.S.; Pesić, S.; Marković, V. Two cases of uveitis masquerade syndrome caused by bilateral intraocular large B-cell lymphoma. Vojnosanit. Pregl. 2013, 70, 1151–1154. [Google Scholar] [CrossRef] [PubMed]
- Shen, K.; Smith, S.V.; Lee, A.G. Acute myelogenous leukemia presenting with uveitis, optic disc edema, and granuloma annulare: Case report. Can. J. Ophthalmol. 2016, 51, e153–e155. [Google Scholar] [CrossRef] [PubMed]
- Miserocchi, E.; Cimminiello, C.; Mazzola, M.; Russo, V.; Modorati, G.M. New-onset uveitis during CTLA-4 blockade therapy with ipilimumab in metastatic melanoma patient. Can. J. Ophthalmol. 2015, 50, e2–e4. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, J.T.; Pasadhika, S.; Crouser, E.D.; Choi, D.; Harrington, C.A.; Lewis, J.A.; Austin, C.R.; Diebel, T.N.; Vance, E.E.; Braziel, R.M.; et al. Hypothesis: Sarcoidosis is a STAT1-mediated disease. Clin. Immunol. 2009, 132, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Yang, Z.; Du, L.; Jiang, Z.; Shu, Q.; Chen, Y.; Li, F.; Zhou, Q.; Ohno, S.; Chen, R.; et al. Identification of a susceptibility locus in STAT4 for Behcet’s disease in Han Chinese in a genome-wide association study. Arthritis. Rheum. 2012, 64, 4104–4113. [Google Scholar] [CrossRef] [PubMed]
- Remmers, E.F.; Cosan, F.; Kirino, Y.; Ombrello, M.J.; Abaci, N.; Satorius, C.; Le, J.M.; Yang, B.; Korman, B.D.; Cakiris, A.; et al. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behcet’s disease. Nat. Genet. 2010, 42, 698–702. [Google Scholar] [CrossRef] [PubMed]
- Mizuki, N.; Meguro, A.; Ota, M.; Ohno, S.; Shiota, T.; Kawagoe, T.; Ito, N.; Kera, J.; Okada, E.; Yatsu, K.; et al. Genome-wide association studies identify IL23R-IL12RB2 and IL10 as Behcet’s disease susceptibility loci. Nat. Genet. 2010, 42, 703–706. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.C.; Claushuis, T.A.; Cortes, A.; Martin, T.M.; Evans, D.M.; Leo, P.; Mukhopadhyay, P.; Bradbury, L.A.; Cremin, K.; Harris, J.; et al. Genetic dissection of acute anterior uveitis reveals similarities and differences in associations observed with ankylosing spondylitis. Arthritis Rheumatol. 2015, 67, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Chen, L.; Tang, J.; Hou, S.; Liao, D.; Ye, Z.; Wang, C.; Cao, Q.; Kijlstra, A.; Yang, P. Association Between Copy Number Variations of TLR7 and Ocular Behcet’s Disease in a Chinese Han Population. Investig. Ophthalmol. Vis. Sci. 2015, 56, 1517–1523. [Google Scholar] [CrossRef] [PubMed]
- Kirino, Y.; Zhou, Q.; Ishigatsubo, Y.; Mizuki, N.; Tugal-Tutkun, I.; Seyahi, E.; Özyazgan, Y.; Ugurlu, S.; Erer, B.; Abaci, N. Targeted resequencing implicates the familial Mediterranean fever gene MEFV and the toll-like receptor 4 gene TLR4 in Behcet disease. Proc. Natl. Acad. Sci. USA 2013, 110, 8134–8139. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Yang, P.; Chu, L.; Zhou, H.; Huang, X.; Zhu, L.; Kijlstra, A. T-bet expression in the iris and spleen parallels disease expression during endotoxin-induced uveitis. Graefe’s Arch. Clin. Exp. Ophthalmol. 2007, 245, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Kirino, Y.; Bertsias, G.; Ishigatsubo, Y.; Mizuki, N.; Tugal-Tutkun, I.; Seyahi, E.; Ozyazgan, Y.; Sacli, F.S.; Erer, B.; Inoko, H.; et al. Genome-wide association analysis identifies new susceptibility loci for Behcet’s disease and epistasis between HLA-B*51 and ERAP1. Nat. Genet. 2013, 45, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Yang, P.; Hou, S.; Du, L.; Xie, L.; Zhou, H.; Kijlstra, A. IL-23R gene confers susceptibility to Behcet’s disease in a Chinese Han population. Ann. Rheum. Dis. 2010, 69, 1325–1328. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.R.; Choi, D.; Chipps, T.J.; Pan, Y.; Zamora, D.O.; Davies, M.H.; Babra, B.; Powers, M.R.; Planck, S.R.; Rosenbaum, J.T. Unique gene expression profiles of donor-matched human retinal and choroidal vascular endothelial cells. Investig. Ophthalmol. Vis. Sci. 2007, 48, 2676–2684. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, B.; Maminishkis, A.; Mahesh, S.P.; Yeh, S.; Lew, J.; Lim, W.K.; Sen, H.N.; Clarke, G.; Buggage, R. Gene expression profiling in autoimmune noninfectious uveitis disease. J. Immunol. 2008, 181, 5147–5157. [Google Scholar] [CrossRef] [PubMed]
- Ohta, K.; Kikuchi, T.; Miyahara, T.; Yoshimura, N. DNA microarray analysis of gene expression in iris and ciliary body of rat eyes with endotoxin-induced uveitis. Exp. Eye Res. 2005, 80, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Mzhesh, S.P.; Liu, B.; Yeh, S.; Lew, J.; Lim, W.; Levy Clarke, G.; Buggage, R.; Nussenblatt, R.B. Gene Expression Profiling of Non-infectious Uveitis Patients Using Pathway Specific cDNA Microarray Analysis. Investig. Ophthalmol. Vis. Sci. 2007, 48, 1505. [Google Scholar] [CrossRef]
- Oliver, S. Guilt-by-association goes global. Nature 2000, 403, 601–603. [Google Scholar] [CrossRef] [PubMed]
- Oti, M.; Snel, B.; Huynen, M.A.; Brunner, H.G. Predicting disease genes using protein-protein interactions. J. Méd. Genet. 2006, 43, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Krauthammer, M.; Kaufmann, C.A.; Conrad Gilliam, T.; Rzhetsky, A. Molecular triangulation: Bridging linkage and molecular-network information for identifying candidate genes in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2004, 101, 15148–15153. [Google Scholar] [CrossRef] [PubMed]
- Kohler, S.; Bauer, S.; Horn, D.; Robinson, P.N. Walking the interactome for prioritization of candidate disease genes. Am. J. Hum. Genet. 2008, 82, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, J. Disease gene identification by random walk on multigraphs merging heterogeneous genomic and phenotype data. BMC Genom. 2012, 13, S27. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Gan, M.X.; He, P. Constructing a gene semantic similarity network for the inference of disease genes. BMC Syst. Biol. 2011, 5, S2. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hao, X.Z.; Huang, T.; Shu, Y.; Huang, G.H.; Li, H.P. Application of the shortest path algorithm for the discovery of breast cancer related genes. Curr. Bioinform. 2016, 11, 51–58. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, J.; Yang, T.; Huang, T.; Shu, Y.; Chen, L. Identification of novel proliferative diabetic retinopathy related genes on protein-protein interaction network. Neurocomputing 2016, 217, 63–72. [Google Scholar] [CrossRef]
- Gui, T.; Dong, X.; Li, R.; Li, Y.; Wang, Z. Identification of Hepatocellular Carcinoma–Related Genes with a Machine Learning and Network Analysis. J. Comput. Biol. 2015, 22, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, J.; Huang, T.; Kong, X.; Lu, L.; Cai, Y.D. Mining for novel tumor suppressor genes using a shortest path approach. J. Biomol. Struct. Dyn. 2016, 34, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Huang, T.; Zhang, Y.H.; Jiang, Y.; Zheng, M.; Cai, Y.D. Identification of novel candidate drivers connecting different dysfunctional levels for lung adenocarcinoma using protein-protein interactions and a shortest path approach. Sci. Rep. 2016, 6, 29849. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, B.; Wang, S.; Yang, J.; Hu, J.; Xie, Z.; Wang, Y.; Huang, T.; Cai, Y.D. OPMSP: A computational method integrating protein interaction and sequence information for the identification of novel putative oncogenes. Protein. Pept. Lett. 2016, 23, 1081–1094. [Google Scholar] [CrossRef] [PubMed]
- Oguz, A.K.; Yılmaz, S.T.; Oygür, Ç.Ş.; Çandar, T.; Sayın, I.; Kılıçoğlu, S.S.; Ergün, İ.; Ateş, A.; Özdağ, H.; Akar, N. Behcet’s: A Disease or a Syndrome? Answer from an Expression Profiling Study. PLoS ONE 2016, 11, e0149052. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, I.; Abe, T.; Narikawa, K.; Feng, J.; Misu, T.; Nakashima, I.; Fujimori, J.; Tamai, M.; Fujihara, K.; Itoyama, Y. Chemokine profile in the cerebrospinal fluid and serum of Vogt-Koyanagi-Harada disease. J. Neuroimmunol. 2005, 158, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.R.; Mahdi, R.R.; Oh, H.M.; Amadi-Obi, A.; Levy-Clarke, G.; Burton, J.; Eseonu, A.; Lee, Y.; Chan, C.C.; Egwuagu, C.E. Suppressor of cytokine signaling-1 (SOCS1) inhibits lymphocyte recruitment into the retina and protects SOCS1 transgenic rats and mice from ocular inflammation. Investig. Ophthalmol. Vis. Sci. 2011, 52, 6978–6986. [Google Scholar] [CrossRef] [PubMed]
- Abu El-Asrar, A.M.; Berghmans, N.; Al-Obeidan, S.A.; Mousa, A.; Opdenakker, G.; van Damme, J.; Struyf, S. The Cytokine Interleukin-6 and the Chemokines CCL20 and CXCL13 Are Novel Biomarkers of Specific Endogenous Uveitic Entities. Investig. Ophthalmol. Vis. Sci. 2016, 57, 4606–4613. [Google Scholar] [CrossRef] [PubMed]
- Hollenbaugh, D.; Grosmaire, L.S.; Kullas, C.D.; Chalupny, N.J.; Braesch-Andersen, S.; Noelle, R.J.; Stamenkovic, I.; Ledbetter, J.A.; Aruffo, A. The human T cell antigen gp39, a member of the TNF gene family, is a ligand for the CD40 receptor: Expression of a soluble form of gp39 with B cell co-stimulatory activity. EMBO J. 1992, 11, 4313–4321. [Google Scholar] [PubMed]
- Lane, P.; Traunecker, A.; Hubele, S.; Inui, S.; Lanzavecchia, A.; Gray, D. Activated human T cells express a ligand for the human B cell-associated antigen CD40 which participates in T cell-dependent activation of B lymphocytes. Eur. J. Immunol. 1992, 22, 2573–2578. [Google Scholar] [CrossRef] [PubMed]
- Noelle, R.J.; Ledbetter, J.A.; Aruffo, A. CD40 and its ligand, an essential ligand-receptor pair for thymus-dependent B-cell activation. Immunol. Today 1992, 13, 431–433. [Google Scholar] [CrossRef]
- Fanslow, W.C.; Srinivasan, S.; Paxton, R.; Gibson, M.G.; Spriggs, M.K.; Armitage, R.J. Structural characteristics of CD40 ligand that determine biological function. Semin. Immunol. 1994, 6, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Howard, L.M.; Miga, A.J.; Vanderlugt, C.L.; Dal Canto, M.C.; Laman, J.D.; Noelle, R.J.; Miller, S.D. Mechanisms of immunotherapeutic intervention by anti-CD40L (CD154) antibody in an animal model of multiple sclerosis. J. Clin. Investig. 1999, 103, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Casamayor-Palleja, M.; Khan, M.; MacLennan, I.C. A subset of CD4+ memory T cells contains preformed CD40 ligand that is rapidly but transiently expressed on their surface after activation through the T cell receptor complex. J. Exp. Med. 1995, 181, 1293–1301. [Google Scholar] [CrossRef] [PubMed]
- Stuber, E.; Strober, W.; Neurath, M. Blocking the CD40L-CD40 interaction in vivo specifically prevents the priming of T helper 1 cells through the inhibition of interleukin 12 secretion. J. Exp. Med. 1996, 183, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Grewal, I.S.; Flavell, R.A. CD40 and CD154 in cell-mediated immunity. Annu. Rev. Immunol. 1998, 16, 111–135. [Google Scholar] [CrossRef] [PubMed]
- Ogard, C.; Sorensen, T.L.; Krogh, E. Increased CD40 ligand in patients with acute anterior uveitis. Acta Ophthalmol. Scand. 2005, 83, 370–373. [Google Scholar] [CrossRef] [PubMed]
- Balashov, K.E.; Smith, D.R.; Khoury, S.J.; Hafler, D.A.; Weiner, H.L. Increased interleukin 12 production in progressive multiple sclerosis: Induction by activated CD4+ T cells via CD40 ligand. Proc. Natl. Acad. Sci. USA 1997, 94, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Ang, M.; Cheung, G.; Vania, M.; Chen, J.; Yang, H.; Li, J.; Chee, S.P. Aqueous cytokine and chemokine analysis in uveitis associated with tuberculosis. Mol. Vis. 2012, 18, 565–573. [Google Scholar] [PubMed]
- Deng, B.; Ye, Z.; Li, L.; Zhang, D.; Zhu, Y.; He, Y.; Wang, C.; Wu, L.; Kijlstra, A.; Yang, P. Higher Expression of NOD1 and NOD2 is Associated with Vogt-Koyanagi-Harada (VKH) Syndrome But Not Behcet’s Disease (BD). Curr. Mol. Med. 2016, 16, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Qi, J.; Wang, Q.; Du, L.; Zhou, Y.; Yu, H.; Kijlstra, A.; Yang, P. Berberine suppresses Th17 and dendritic cell responses. Investig. Ophthalmol. Vis. Sci. 2013, 54, 2516–2522. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.Y.; Wu, W.J.; Huang, C.N.; Li, C.C.; Li, W.M.; Yeh, B.W.; Liang, P.I.; Wu, T.F.; Li, C.F. CSF2 Overexpression Is Associated with STAT5 Phosphorylation and Poor Prognosis in Patients with Urothelial Carcinoma. J. Cancer 2016, 7, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Jucker, M.; Feldman, R.A. Identification of a new adapter protein that may link the common β subunit of the receptor for granulocyte/macrophage colony-stimulating factor, interleukin (IL)-3, and IL-5 to phosphatidylinositol 3-kinase. J. Biol. Chem. 1995, 270, 27817–27822. [Google Scholar] [PubMed]
- Bittorf, T.; Jaster, R.; Brock, J. Rapid activation of the MAP kinase pathway in hematopoietic cells by erythropoietin, granulocyte-macrophage colony-stimulating factor and interleukin-3. Cell Signal. 1994, 6, 305–311. [Google Scholar] [CrossRef]
- Kimura, A.; Rieger, M.A.; Simone, J.M.; Chen, W.; Wickre, M.C.; Zhu, B.M.; Hoppe, P.S.; O’Shea, J.J.; Schroeder, T.; Hennighausen, L. The transcription factors STAT5A/B regulate GM-CSF-mediated granulopoiesis. Blood 2009, 114, 4721–4728. [Google Scholar] [CrossRef] [PubMed]
- Mui, A.L.; Wakao, H.; Harada, N.; O’Farrell, A.M.; Miyajima, A. Interleukin-3, granulocyte-macrophage colony-stimulating factor, and interleukin-5 transduce signals through two forms of STAT5. J. Leukoc. Biol. 1995, 57, 799–803. [Google Scholar] [PubMed]
- Feldman, G.M.; Rosenthal, L.A.; Liu, X.; Hayes, M.P.; Wynshaw-Boris, A.; Leonard, W.J.; Hennighausen, L.; Finbloom, D.S. STAT5A-deficient mice demonstrate a defect in granulocyte-macrophage colony-stimulating factor-induced proliferation and gene expression. Blood 1997, 90, 1768–1776. [Google Scholar] [PubMed]
- Taheri, S.; Borlu, M.; Evereklioglu, C.; Ozdemir, S.Y.; Ozkul, Y. mRNA Expression Level of Interleukin Genes in the Determining Phases of Behcet’s Disease. Ann. Dermatol. 2015, 27, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Tan, X.; Zhou, Q.; Zhu, Y.; Tian, Y.; Yu, H.; Kijlstra, A.; Yang, P. IL-1β triggered by peptidoglycan and lipopolysaccharide through TLR2/4 and ROS-NLRP3 inflammasome-dependent pathways is involved in ocular Behcet’s disease. Investig. Ophthalmol. Vis. Sci. 2013, 54, 402–414. [Google Scholar] [CrossRef] [PubMed]
- Roberge, F.G.; de Smet, M.D.; Benichou, J.; Kriete, M.F.; Raber, J.; Hakimi, J. Treatment of uveitis with recombinant human interleukin-13. Br. J. Ophthalmol. 1998, 82, 1195–1198. [Google Scholar] [CrossRef] [PubMed]
- Marie, O.; Thillaye-Goldenberg, B.; Naud, M.C.; de Kozak, Y. Inhibition of endotoxin-induced uveitis and potentiation of local TNF-α and interleukin-6 mRNA expression by interleukin-13. Investig. Ophthalmol. Vis. Sci. 1999, 40, 2275–2282. [Google Scholar]
- Lemaitre, C.; Thillaye-Goldenberg, B.; Naud, M.C.; de Kozak, Y. The effects of intraocular injection of interleukin-13 on endotoxin-induced uveitis in rats. Investig. Ophthalmol. Vis. Sci. 2001, 42, 2022–2030. [Google Scholar]
- De Kozak, Y.; Omri, B.; Smith, J.R.; Naud, M.C.; Thillaye-Goldenberg, B.; Crisanti, P. Protein kinase C ζ (PKC ζ) regulates ocular inflammation and apoptosis in endotoxin-induced uveitis (EIU)—Signaling molecules involved in EIU resolution by PKC ζ inhibitor and interleukin-13. Am. J. Pathol. 2007, 170, 1241–1257. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.J.; Kong, X.L.; Zhang, P. [In vivo effect of recombined IL-15/Fc fusion protein on EAU]. Sichuan Da Xue Xue Bao Yi Xue Ban 2008, 39, 944–949. [Google Scholar] [PubMed]
- Choe, J.Y.; Lee, H.; Kim, S.G.; Kim, M.J.; Park, S.H.; Kim, S.K. The distinct expressions of interleukin-15 and interleukin-15 receptor α in Behcet’s disease. Rheumatol. Int. 2013, 33, 2109–2115. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Liao, D.; Zhang, J.; Fang, J.; Chen, L.; Qi, J.; Zhang, Q.; Liu, Y.; Bai, L.; Zhou, Y.; et al. Genetic variations of IL17F and IL23A show associations with Behcet’s disease and Vogt-Koyanagi-Harada syndrome. Ophthalmology 2015, 122, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Lim, W.K.; Fujimoto, C.; Ursea, R.; Mahesh, S.P.; Silver, P.; Chan, C.C.; Gery, I.; Nussenblatt, R.B. Suppression of immune-mediated ocular inflammation in mice by interleukin 1 receptor antagonist administration. Arch. Ophthalmol. 2005, 123, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Lindner, E.; Weger, M.; Steinwender, G.; Griesbacher, A.; Posch, U.; Ulrich, S.; Wegscheider, B.; Ardjomand, N.; El-Shabrawi, Y. IL2RA gene polymorphism rs2104286 A>G seen in multiple sclerosis is associated with intermediate uveitis: Possible parallel pathways? Investig. Ophthalmol. Vis. Sci. 2011, 52, 8295–8299. [Google Scholar] [CrossRef] [PubMed]
- Nussenblatt, R.B.; Fortin, E.; Schiffman, R.; Rizzo, L.; Smith, J.; van Veldhuisen, P.; Sran, P.; Yaffe, A.; Goldman, C.K.; Waldmann, T.A.; et al. Treatment of noninfectious intermediate and posterior uveitis with the humanized anti-Tac mAb: A phase I/II clinical trial. Proc. Natl. Acad. Sci. USA 1999, 96, 7462–7466. [Google Scholar] [CrossRef] [PubMed]
- Cenit, M.C.; Marquez, A.; Cordero-Coma, M.; Fonollosa, A.; Adan, A.; Martinez-Berriotxoa, A.; Llorenc, V.; Diaz Valle, D.; Blanco, R.; Canal, J.; et al. Evaluation of the IL2/IL21, IL2RA and IL2RB genetic variants influence on the endogenous non-anterior uveitis genetic predisposition. BMC Med. Genet. 2013, 14, 52. [Google Scholar] [CrossRef] [PubMed]
- Niven, J.; Hoare, J.; McGowan, D.; Devarajan, G.; Itohara, S.; Gannage, M.; Teismann, P.; Crane, I. S100B Up-Regulates Macrophage Production of IL1β and CCL22 and Influences Severity of Retinal Inflammation. PLoS ONE 2015, 10, e0132688. [Google Scholar] [CrossRef] [PubMed]
- Charteris, D.G.; Lightman, S.L. Comparison of the expression of interferon gamma, IL2, IL4, and lymphotoxin mRNA in experimental autoimmune uveoretinitis. Br. J. Ophthalmol. 1994, 78, 786–790. [Google Scholar] [CrossRef] [PubMed]
- Shahram, F.; Nikoopour, E.; Rezaei, N.; Saeedfar, K.; Ziaei, N.; Davatchi, F.; Amirzargar, A. Association of interleukin-2, interleukin-4 and transforming growth factor-β gene polymorphisms with Behcet’s disease. Clin. Exp. Rheumatol. 2011, 29, S28–S31. [Google Scholar] [PubMed]
- Chang, J.H.; McCluskey, P.; Wakefield, D. Expression of toll-like receptor 4 and its associated lipopolysaccharide receptor complex by resident antigen-presenting cells in the human uvea. Investig. Ophthalmol. Vis. Sci. 2004, 45, 1871–1878. [Google Scholar] [CrossRef]
- Chang, J.H.; McCluskey, P.J.; Wakefield, D. Toll-like receptors in ocular immunity and the immunopathogenesis of inflammatory eye disease. Br. J. Ophthalmol. 2006, 90, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Meguro, A.; Ota, M.; Katsuyama, Y.; Oka, A.; Ohno, S.; Inoko, H.; Mizuki, N. Association of the toll-like receptor 4 gene polymorphisms with Behcet’s disease. Ann. Rheum. Dis. 2008, 67, 725–727. [Google Scholar] [CrossRef] [PubMed]
- Song, G.G.; Choi, S.J.; Ji, J.D.; Lee, Y.H. Toll-like receptor polymorphisms and vasculitis susceptibility: Meta-analysis and systematic review. Mol. Biol. Rep. 2013, 40, 1315–1323. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Hu, R.; Hou, S.; Ye, Z.; Xiang, Q.; Qi, J.; Zhou, Y.; Kijlstra, A.; Yang, P. Association of TLR2 gene polymorphisms with ocular Behcet’s disease in a Chinese Han population. Investig. Ophthalmol. Vis. Sci. 2013, 54, 8384–8392. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Tan, X.; Zhou, Q.; Tian, Y.; Kijlstra, A.; Yang, P. TLR3 and TLR4 But not TLR2 are Involved in Vogt-Koyanagi-Harada Disease by Triggering Proinflammatory Cytokines Production Through Promoting the Production of Mitochondrial Reactive Oxygen Species. Curr. Mol. Med. 2015, 15, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yan, H.; Sun, B.; Zuo, A.; Liang, D. Subretinal transfection of chitosan-loaded TLR3-siRNA for the treatment of experimental autoimmune uveitis. Eur. J. Pharm. Biopharm. 2013, 85, 726–735. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Chen, L.; Tang, J.H.; Hou, S.P.; Liao, D.; Ye, Z.; Wang, C.K.; Cao, Q.F.; Kijlstra, A.; Yang, P.Z. Association Between Copy Number Variations of TLR7 and Ocular Behcet’s Disease in a Chinese Han Population. Investig. Ophthalmol. Vis. Sci. 2015, 56, 1517–1523. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, N.; Sekine, H.; Kobayashi, H.; Sato, Y.; Ohira, H. Association of the toll-like receptor 9 gene polymorphisms with Behcet’s disease in a Japanese population. Fukushima J. Med. Sci. 2012, 58, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.P.; Pei, Y.X.; Li, G.F.; Lou, Y.R. Effect of glucocorticoid on cytokines TLR9 and TLR7 in peripheral blood for patients with uveitis. Exp. Ther. Med. 2016, 12, 3893–3896. [Google Scholar] [CrossRef] [PubMed]
- Shaulian, E. AP-1—The Jun proteins: Oncogenes or tumor suppressors in disguise? Cell Signal. 2010, 22, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Smeal, T.; Binetruy, B.; Mercola, D.; Grover-Bardwick, A.; Heidecker, G.; Rapp, U.R.; Karin, M. Oncoprotein-mediated signalling cascade stimulates c-Jun activity by phosphorylation of serines 63 and 73. Mol. Cell Biol. 1992, 12, 3507–3513. [Google Scholar] [CrossRef] [PubMed]
- Pulverer, B.J.; Kyriakis, J.M.; Avruch, J.; Nikolakaki, E.; Woodgett, J.R. Phosphorylation of c-jun mediated by MAP kinases. Nature 1991, 353, 670–674. [Google Scholar] [CrossRef] [PubMed]
- Qing, H.; Gong, W.; Che, Y.; Wang, X.; Peng, L.; Liang, Y.; Wang, W.; Deng, Q.; Zhang, H.; Jiang, B. PAK1-dependent MAPK pathway activation is required for colorectal cancer cell proliferation. Tumour Biol. 2012, 33, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Chen, C.L.; Tseng, Y.L.; Fang, Y.T.; Lin, Y.S.; Su, W.C.; Chen, C.C.; Chang, K.C.; Wang, Y.C.; Lin, C.F. Annexin A2 silencing induces G2 arrest of non-small cell lung cancer cells through p53-dependent and -independent mechanisms. J. Biol. Chem. 2012, 287, 32512–32524. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Villasana, V.; Gutierrez-Puente, Y.; Tari, A.M. Cyclooxygenase-2 utilizes Jun N-terminal kinases to induce invasion, but not tamoxifen resistance, in MCF-7 breast cancer cells. Oncol. Rep. 2013, 30, 1506–1510. [Google Scholar] [PubMed]
- Gao, L.; Huang, S.; Ren, W.; Zhao, L.; Li, J.; Zhi, K.; Zhang, Y.; Qi, H.; Huang, C. Jun activation domain-binding protein 1 expression in oral squamous cell carcinomas inversely correlates with the cell cycle inhibitor p27. Med. Oncol. 2012, 29, 2499–2504. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Tao, Y.G.; Deng, X.Y.; Jin, X.; Tan, Y.N.; Tang, M.; Wu, Q.; Lee, L.M.; Cao, Y. Heterodimer formation between c-Jun and Jun B proteins mediated by Epstein-Barr virus encoded latent membrane protein 1. Cell Signal. 2004, 16, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Rocha, G.; Duclos, A.; Chalifour, L.E.; Baines, M.G.; Antecka, E.; Deschenes, J. Analysis of gene expression during experimental uveitis in the rabbit. Can. J. Ophthalmol. 1996, 31, 228–233. [Google Scholar] [PubMed]
- Turner, M.J.; DaSilva-Arnold, S.; Luo, N.; Hu, X.; West, C.C.; Sun, L.; Hall, C.; Bradish, J.; Kaplan, M.H.; Travers, J.B.; et al. STAT6-mediated keratitis and blepharitis: A novel murine model of ocular atopic dermatitis. Investig. Ophthalmol. Vis. Sci. 2014, 55, 3803–3808. [Google Scholar] [CrossRef] [PubMed]
- Tepper, R.I.; Levinson, D.A.; Stanger, B.Z.; Campos-Torres, J.; Abbas, A.K.; Leder, P. IL-4 induces allergic-like inflammatory disease and alters T cell development in transgenic mice. Cell 1990, 62, 457–467. [Google Scholar] [CrossRef]
- Amadi-Obi, A.; Yu, C.R.; Liu, X.; Mahdi, R.M.; Clarke, G.L.; Nussenblatt, R.B.; Gery, I.; Lee, Y.S.; Egwuagu, C.E. TH17 cells contribute to uveitis and scleritis and are expanded by IL-2 and inhibited by IL-27/STAT1. Nat. Med. 2007, 13, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Malla, N.; Sjoli, S.; Winberg, J.O.; Hadler-Olsen, E.; Uhlin-Hansen, L. Biological and pathobiological functions of gelatinase dimers and complexes. Connect. Tissue Res. 2008, 49, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Murphy, G.; Nagase, H. Progress in matrix metalloproteinase research. Mol. Asp. Med. 2008, 29, 290–308. [Google Scholar] [CrossRef] [PubMed]
- Sivak, J.M.; Fini, M.E. Mmps in the eye: Emerging roles for matrix metalloproteinases in ocular physiology. Prog. Retin. Eye Res. 2002, 21, 1–14. [Google Scholar] [CrossRef]
- Nagase, H.; Visse, R.; Murphy, G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res. 2006, 69, 562–573. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Kang, S.W.; Baek, H.J.; Choi, H.J.; Bae, Y.D.; Kang, E.H.; Lee, E.Y.; Lee, E.B.; Song, Y.W. Association between matrix metalloproteinase 9 promoter polymorphisms and Behcet’s disease. Hum. Immunol. 2010, 71, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Quillard, T.; Coupel, S.; Coulon, F.; Fitau, J.; Chatelais, M.; Cuturi, M.C.; Chiffoleau, E.; Charreau, B. Impaired Notch4 activity elicits endothelial cell activation and apoptosis: Implication for transplant arteriosclerosis. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 2258–2265. [Google Scholar] [CrossRef] [PubMed]
- Verginelli, F.; Adesso, L.; Limon, I.; Alisi, A.; Gueguen, M.; Panera, N.; Giorda, E.; Raimondi, L.; Ciarapica, R.; Campese, A.F.; et al. Activation of an endothelial Notch1-Jagged1 circuit induces VCAM1 expression, an effect amplified by interleukin-1β. Oncotarget 2015, 6, 43216–43229. [Google Scholar] [PubMed]
- Crosson, J.N.; Laird, P.W.; Debiec, M.; Bergstrom, C.S.; Lawson, D.H.; Yeh, S. Vogt-Koyanagi-Harada-like syndrome after CTLA-4 inhibition with ipilimumab for metastatic melanoma. J. Immunother. 2015, 38, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.R.; Kim, S.H.; Mahdi, R.M.; Egwuagu, C.E. SOCS3 deletion in T lymphocytes suppresses development of chronic ocular inflammation via upregulation of CTLA-4 and expansion of regulatory T cells. J. Immunol. 2013, 191, 5036–5043. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, J.; Izumi, T.; Arimitsu, N.; Fujiwara, N.; Ueda, Y.; Wakisaka, S.; Yoshikawa, H.; Kaneko, F.; Suzuki, T.; Takai, K.; et al. Skewed TGFβ/Smad signalling pathway in T cells in patients with Behcet’s disease. Clin. Exp. Rheumatol. 2012, 30, S35–S39. [Google Scholar] [PubMed]
- Li, Q.; Sun, B.; Dastgheib, K.; Chan, C.C. Suppressive effect of transforming growth factor β1 on the recurrence of experimental melanin protein-induced uveitis: Upregulation of ocular interleukin-10. Clin. Immunol. Immunopathol. 1996, 81, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.K.; Gupta, A.; Kamal, S.; Bansal, R.; Singh, N.; Sharma, K.; Virk, S.; Sachdeva, N. Role of Regulatory T Cells in Tubercular Uveitis. Ocul. Immunol. Inflamm. 2016, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fabiani, C.; Vitale, A.; Lopalco, G.; Iannone, F.; Frediani, B.; Cantarini, L. Different roles of TNF inhibitors in acute anterior uveitis associated with ankylosing spondylitis: State of the art. Clin. Rheumatol. 2016, 35, 2631–2638. [Google Scholar] [CrossRef] [PubMed]
- Hatemi, I.; Hatemi, G.; Pamuk, O.N.; Erzin, Y.; Celik, A.F. TNF-α antagonists and thalidomide for the management of gastrointestinal Behcet’s syndrome refractory to the conventional treatment modalities: A case series and review of the literature. Clin. Exp. Rheumatol. 2015, 33, S129–S137. [Google Scholar] [PubMed]
- Bharadwaj, A.S.; Schewitz-Bowers, L.P.; Wei, L.; Lee, R.W.; Smith, J.R. Intercellular adhesion molecule 1 mediates migration of Th1 and Th17 cells across human retinal vascular endothelium. Investig. Ophthalmol. Vis. Sci. 2013, 54, 6917–6925. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Huang, T.; Shi, X.; Lu, W.C.; Cai, Y.D.; Chou, K.C. Predicting functions of proteins in mouse based on weighted protein-protein interaction network and protein hybrid properties. PLoS ONE 2011, 6, e14556. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.L.; Huang, T.; Cai, Y.D.; Chou, K.C. Prediction of Body Fluids where Proteins are Secreted into Based on Protein Interaction Network. PLoS ONE 2011, 6, e22989. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, Y.H.; Huang, T.; Cai, Y.D. Identifying novel protein phenotype annotations by hybridizing protein-protein interactions and protein sequence similarities. Mol. Genet. Genom. 2016, 291, 913–934. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.J.; Kuhn, M.; Stark, M.; Chaffron, S.; Creevey, C.; Muller, J.; Doerks, T.; Julien, P.; Roth, A.; Simonovic, M. STRING 8-a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009, 37, D412–D416. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Patra, J.C. Genome-wide inferring genephenotype relationship by walking on the heterogeneous network. Bioinformatics 2010, 26, 1219–1224. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, L.; Kong, X.; Huang, T.; Cai, Y.-D. Analysis of Tumor Suppressor Genes Based on Gene Ontology and the KEGG Pathway. PLoS ONE 2014, 9, e107202. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Zhang, J.; Xu, Z.P.; Hu, L.L.; Chen, L.; Shao, J.L.; Zhang, L.; Kong, X.Y.; Cai, Y.D.; Chou, K.C. Deciphering the effects of gene deletion on yeast longevity using network and machine learning approaches. Biochimie 2012, 94, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xing, Z.; Ma, M.; Wang, N.; Cai, Y.-D.; Chen, L.; Xu, X. Gene Ontology and KEGG Enrichment Analyses of Genes Related to Age-Related Macular Degeneration. BioMed Res. Int. 2014, 2014, 450386. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, Y.-H.; Zheng, M.; Huang, T.; Cai, Y.-D. Identification of compound-protein interactions through the analysis of gene ontology, KEGG enrichment for proteins and molecular fragments of compounds. Mol. Genet. Genom. 2016, 291, 2065–2079. [Google Scholar] [CrossRef] [PubMed]
- Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In Proceedings of the 14th International joint Conference on artificial intelligence, Montreal, QC, Canada, 20–25 August 1995. [Google Scholar]
| Index of Part | Recall | Precision | F1-Measure |
|---|---|---|---|
| 1 | 0.172 | 0.089 | 0.118 |
| 2 | 0.172 | 0.088 | 0.116 |
| 3 | 0.379 | 0.177 | 0.242 |
| 4 | 0.310 | 0.141 | 0.194 |
| 5 | 0.400 | 0.211 | 0.276 |
| Mean | 0.287 | 0.141 | 0.189 |
| Ensembl ID | Gene Symbol | Description | Probability | p-Value | MIS | MFS |
|---|---|---|---|---|---|---|
| ENSP00000351671 b | CCL20 | C-C motif chemokine ligand 20 | 1.65 × 10−4 | <0.001 | 999 | 0.841 |
| ENSP00000250151 b | CCL4 | C-C motif chemokine ligand 4 | 1.64 × 10−4 | <0.001 | 994 | 0.820 |
| ENSP00000326432 c | CCR8 | C-C motif chemokine receptor 8 | 8.90 × 10−5 | <0.001 | 951 | 0.816 |
| ENSP00000313419 b | CD19 | CD19 molecule | 2.15 × 10−4 | <0.001 | 947 | 0.837 |
| ENSP00000320084 c | CD276 | CD276 molecule | 1.91 × 10−4 | <0.001 | 955 | 0.823 |
| ENSP00000359663 b | CD40LG | CD40 ligand | 1.97 × 10−4 | <0.001 | 999 | 0.839 |
| ENSP00000264246 b | CD80 | CD80 molecule | 2.18 × 10−4 | <0.001 | 999 | 0.820 |
| ENSP00000283635 c | CD8A | CD8a molecule | 1.91 × 10−4 | <0.001 | 990 | 0.815 |
| ENSP00000296871 c | CSF2 | Colony stimulating factor 2 | 2.71 × 10−4 | <0.001 | 992 | 0.875 |
| ENSP00000225474 c | CSF3 | Colony stimulating factor 3 | 1.55 × 10−4 | <0.001 | 916 | 0.829 |
| ENSP00000379110 b | CXCL1 | C-X-C motif chemokine ligand 1 | 1.69 × 10−4 | <0.001 | 973 | 0.827 |
| ENSP00000306884 b | CXCL11 | C-X-C motif chemokine ligand 11 | 1.28 × 10−4 | <0.001 | 999 | 0.818 |
| ENSP00000286758 b | CXCL13 | C-X-C motif chemokine ligand 13 | 1.49 × 10−4 | <0.001 | 986 | 0.806 |
| ENSP00000293778 b | CXCL16 | C-X-C motif chemokine ligand 16 | 1.02 × 10−4 | <0.001 | 952 | 0.800 |
| ENSP00000296027 b | CXCL5 | C-X-C motif chemokine ligand 5 | 1.11 × 10−4 | <0.001 | 958 | 0.811 |
| ENSP00000354901 b | CXCL9 | C-X-C motif chemokine ligand 9 | 2.13 × 10−4 | <0.001 | 999 | 0.883 |
| ENSP00000295683 c | CXCR1 | C-X-C motif chemokine receptor 1 | 8.67 × 10−5 | <0.001 | 999 | 0.833 |
| ENSP00000319635 b | CXCR2 | C-X-C motif chemokine receptor 2 | 1.02 × 10−4 | <0.001 | 999 | 0.851 |
| ENSP00000229239 c | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 2.12 × 10−4 | <0.001 | 922 | 0.824 |
| ENSP00000216341 c | GZMB | Granzyme B | 2.46 × 10−4 | <0.001 | 991 | 0.829 |
| ENSP00000364114 c | HLA-DRB5 | Major histocompatibility complex, class II, DR β 5 | 2.27 × 10−4 | <0.001 | 948 | 0.822 |
| ENSP00000304915 a | IL13 | Interleukin 13 | 1.31 × 10−4 | <0.001 | 999 | 0.813 |
| ENSP00000296545 b | IL15 | Interleukin 15 | 1.85 × 10−4 | <0.001 | 946 | 0.806 |
| ENSP00000263339 b | IL1A | Interleukin 1 α | 1.82 × 10−4 | <0.001 | 996 | 0.820 |
| ENSP00000263341 b | IL1B | Interleukin 1 β | 3.58 × 10−4 | <0.001 | 999 | 0.873 |
| ENSP00000259206 a | IL1RN | Interleukin 1 receptor antagonist | 1.68 × 10−4 | <0.001 | 999 | 0.836 |
| ENSP00000228534 b | IL23A | Interleukin 23 subunit Α | 2.87 × 10−4 | <0.001 | 998 | 0.844 |
| ENSP00000369293 b | IL2RA | Interleukin 2 receptor subunit Α | 2.46 × 10−4 | <0.001 | 999 | 0.866 |
| ENSP00000274520 c | IL9 | Interleukin 9 | 1.27 × 10−4 | <0.001 | 965 | 0.806 |
| ENSP00000360266 b | JUN | Jun proto-oncogene, AP-1 transcription factor subunit | 3.22 × 10−4 | <0.001 | 999 | 0.831 |
| ENSP00000361405 b | MMP9 | Matrix metallopeptidase 9 | 1.70 × 10−4 | <0.001 | 971 | 0.833 |
| ENSP00000379625 a | MYD88 | Myeloid differentiation primary response 88 | 1.82 × 10−4 | <0.001 | 999 | 0.882 |
| ENSP00000356346 c | PTPRC | Protein tyrosine phosphatase, receptor type C | 2.18 × 10−4 | <0.001 | 994 | 0.826 |
| ENSP00000331736 c | SELE | Selectin E | 1.46 × 10−4 | <0.001 | 978 | 0.830 |
| ENSP00000354394 b | STAT1 | Signal transducer and activator of transcription 1 | 2.63 × 10−4 | <0.001 | 999 | 0.852 |
| ENSP00000300134 b | STAT6 | Signal transducer and activator of transcription 6 | 1.77 × 10−4 | <0.001 | 999 | 0.804 |
| ENSP00000221930 a | TGFB1 | Transforming growth factor β 1 | 2.90 × 10−4 | <0.001 | 997 | 0.832 |
| ENSP00000416330 c | TGFBI | Transforming growth factor β induced | 1.91 × 10−4 | <0.001 | 917 | 0.813 |
| ENSP00000260010 b | TLR2 | Toll like receptor 2 | 2.25 × 10−4 | <0.001 | 968 | 0.888 |
| ENSP00000370034 b | TLR7 | Toll like receptor 7 | 1.26 × 10−4 | <0.001 | 926 | 0.819 |
| ENSP00000353874 b | TLR9 | Toll like receptor 9 | 1.55 × 10−4 | <0.001 | 958 | 0.854 |
| ENSP00000294728 b | VCAM1 | Vascular cell adhesion molecule 1 | 2.23 × 10−4 | <0.001 | 968 | 0.882 |
| ENSP00000292174 c | CXCR5 | C-X-C motif chemokine receptor 5 | 1.14 × 10−4 | 0.001 | 976 | 0.820 |
| ENSP00000343204 a | JAK1 | Janus kinase 1 | 1.21 × 10−4 | 0.001 | 999 | 0.818 |
| ENSP00000162749 b | TNFRSF1A | TNF Receptor superfamily member 1A | 2.30 × 10−4 | 0.001 | 999 | 0.826 |
| ENSP00000304414 c | CXCR6 | C-X-C motif chemokine receptor 6 | 9.27 × 10−5 | 0.002 | 964 | 0.803 |
| ENSP00000296795 a | TLR3 | Toll like receptor 3 | 1.58 × 10−4 | 0.002 | 966 | 0.858 |
| ENSP00000231454 c | IL5 | Interleukin 5 | 1.13 × 10−4 | 0.004 | 991 | 0.803 |
| ENSP00000222823 a | NOD1 | Nucleotide binding oligomerization domain containing 1 | 7.72 × 10−5 | 0.004 | 991 | 0.866 |
| ENSP00000231449 b | IL4 | Interleukin 4 | 2.55 × 10−4 | 0.005 | 999 | 0.852 |
| ENSP00000356438 a | PTGS2 | Prostaglandin-endoperoxide synthase 2 | 1.92 × 10−4 | 0.009 | 972 | 0.864 |
| ENSP00000219244 b | CCL17 | C-C motif chemokine ligand 17 | 1.20 × 10−4 | 0.01 | 984 | 0.808 |
| ENSP00000351273 b | CASP8 | Caspase 8 | 9.66 × 10−5 | 0.027 | 999 | 0.821 |
| ENSP00000353483 c | MAPK8 | Mitogen-activated protein kinase 8 | 1.03 × 10−4 | 0.034 | 925 | 0.847 |
| ENSP00000228280 c | KITLG | KIT ligand | 9.60 × 10−5 | 0.039 | 958 | 0.810 |
| ENSP00000238682 c | TGFB3 | Transforming growth factor β 3 | 5.37 × 10−5 | 0.049 | 961 | 0.850 |
| Index of Part | RWR-Based Method | GBA-Based Method | |||||
|---|---|---|---|---|---|---|---|
| Recall | Precision | F1-Measure | Best Value of k | Recall | Precision | F1-Measure | |
| 1 | 0.172 | 0.089 | 0.118 | 1 | 0.207 | 0.061 | 0.094 |
| 2 | 0.172 | 0.088 | 0.116 | 1 | 0.207 | 0.059 | 0.092 |
| 3 | 0.379 | 0.177 | 0.242 | 3 | 0.345 | 0.039 | 0.069 |
| 4 | 0.310 | 0.141 | 0.194 | 1 | 0.172 | 0.052 | 0.079 |
| 5 | 0.400 | 0.211 | 0.276 | 3 | 0.500 | 0.061 | 0.109 |
| RWR-Based Method |
|---|
| Input: Ensembl IDs of uveitis-related genes, a PPI network Output: A number of putative uveitis-related genes |
|
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, S.; Yan, Y.; Li, Z.; Chen, L.; Yang, J.; Zhang, Y.; Wang, S.; Liu, L. Determination of Genes Related to Uveitis by Utilization of the Random Walk with Restart Algorithm on a Protein–Protein Interaction Network. Int. J. Mol. Sci. 2017, 18, 1045. https://doi.org/10.3390/ijms18051045
Lu S, Yan Y, Li Z, Chen L, Yang J, Zhang Y, Wang S, Liu L. Determination of Genes Related to Uveitis by Utilization of the Random Walk with Restart Algorithm on a Protein–Protein Interaction Network. International Journal of Molecular Sciences. 2017; 18(5):1045. https://doi.org/10.3390/ijms18051045
Chicago/Turabian StyleLu, Shiheng, Yan Yan, Zhen Li, Lei Chen, Jing Yang, Yuhang Zhang, Shaopeng Wang, and Lin Liu. 2017. "Determination of Genes Related to Uveitis by Utilization of the Random Walk with Restart Algorithm on a Protein–Protein Interaction Network" International Journal of Molecular Sciences 18, no. 5: 1045. https://doi.org/10.3390/ijms18051045