Long-Term Follow-Up of Resistance-Associated Substitutions in Hepatitis C Virus in Patients in Which Direct Acting Antiviral-Based Therapy Failed
Abstract
:1. Introduction
2. Results
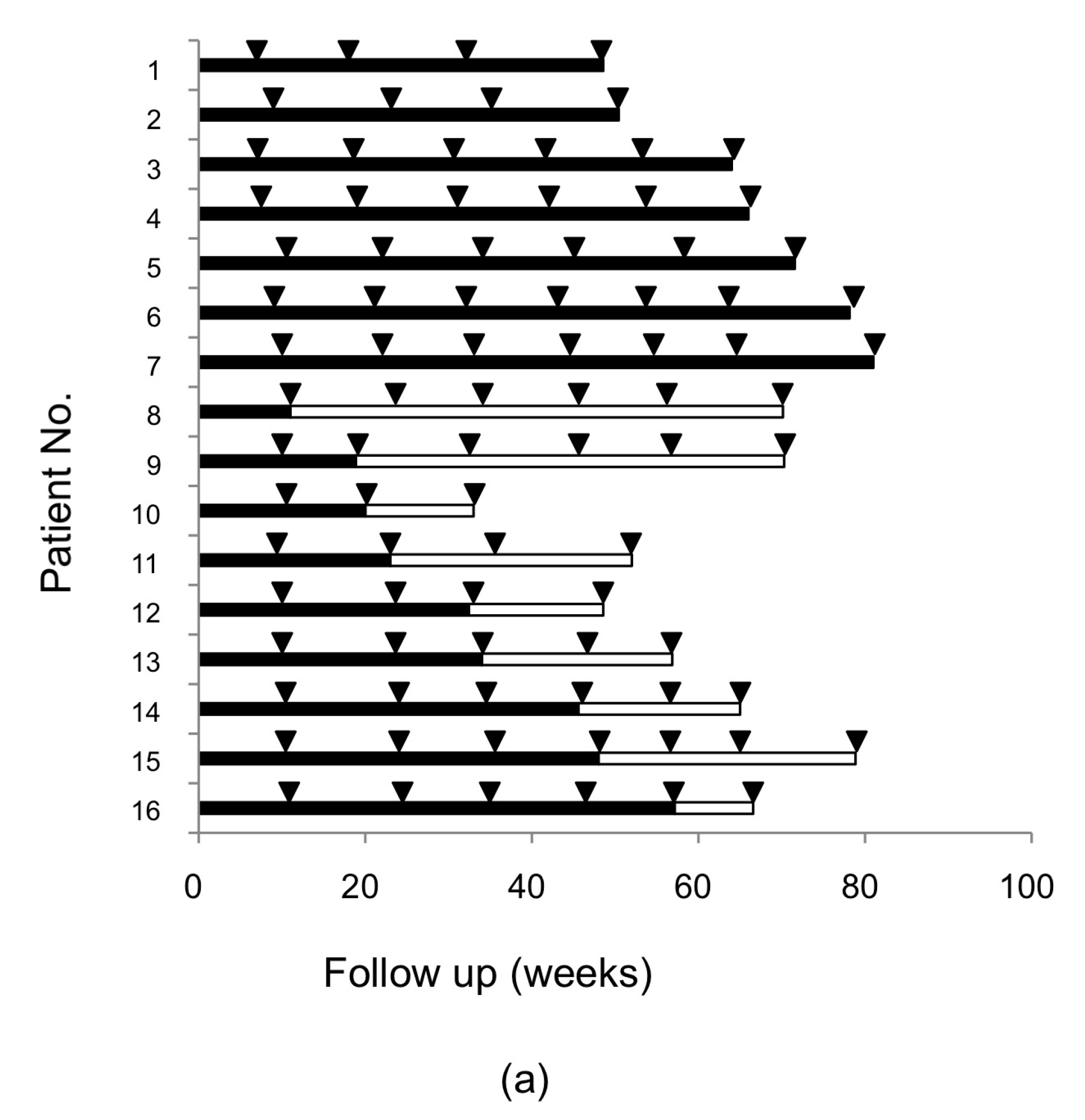
2.1. Resistance-Associated Substitutions (RASs) in the NS3/4A, NS5A, and NS5B Regions of HCV after SMV/PEG-IFN/RBV Treatment
2.2. RASs in the NS3/4A, NS5A, and NS5B Regions of Hepatitis C Virus (HCV) after Daclatasvir/Asunaprevir (DCV/ASV) Treatment
2.3. Alteration of RASs at D168 in the HCV NS3/4A Region and at Y93 in the NS5A Region in Patients Who Failed DCV/ASV Treatment
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. RNA Extraction, cDNA Synthesis, and Direct Sequencing of the NS3/4A, NS5A, and NS5B Regions of the HCV Genome
4.3. Quantitative Y93H Assay of the NS5A Region of the HCV Genome
4.4. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| RAS | Resistance-associated substitution |
| DAA | Direct-acting antiviral |
| SMV/PEG-IFN/RBV | Simeprevir/pegylated-interferon/ribavirin |
| DCV/ASV | Daclatasvir/asunaprevir |
| CHC | Chronic hepatitis C |
| HCV | Hepatitis C virus |
| HCC | Hepatocellular carcinoma |
| SVR | Sustained viral response |
References
- Shepard, C.W.; Finelli, L.; Alter, M.J. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 2005, 5, 558–567. [Google Scholar] [CrossRef]
- Fried, M.W.; Buti, M.; Dore, G.J.; Flisiak, R.; Ferenci, P.; Jacobson, I.; Marcellin, P.; Manns, M.; Nikitin, I.; Poordad, F.; et al. Once-daily simeprevir (TMC435) with pegylated interferon and ribavirin in treatment-naive genotype 1 hepatitis C: The randomized PILLAR study. Hepatology 2013, 58, 1918–1929. [Google Scholar] [CrossRef] [PubMed]
- Manns, M.P.; Gane, E.; Rodriguez-Torres, M.; Stoehr, A.; Yeh, C.T.; Marcellin, P.; Wiedmann, R.T.; Hwang, P.M.; Caro, L.; Barnard, R.J.; et al. Vaniprevir with pegylated interferon α-2a and ribavirin in treatment-naive patients with chronic hepatitis C: A randomized phase II study. Hepatology 2012, 56, 884–893. [Google Scholar] [CrossRef] [PubMed]
- McHutchison, J.G.; Everson, G.T.; Gordon, S.C.; Jacobson, I.M.; Sulkowski, M.; Kauffman, R.; McNair, L.; Alam, J.; Muir, A.J.; Team, P.S. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N. Engl. J. Med. 2009, 360, 1827–1838. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Yasui, S.; Nakamura, M.; Suzuki, E.; Arai, M.; Haga, Y.; Sasaki, R.; Wu, S.; Nakamoto, S.; Imazeki, F.; et al. Daclatasvir plus asunaprevir treatment for real-world HCV genotype 1-infected patients in Japan. Int. J. Med. Sci. 2016, 13, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Tamori, A.; Yoshida, K.; Kurai, O.; Kioka, K.; Hai, H.; Kozuka, R.; Motoyama, H.; Kawamura, E.; Hagihara, A.; Uchida-Kobayashi, S.; et al. Randomized trial of combined triple therapy comprising two types of peginterferon with simeprevir in patients with hepatitis C virus genotype 1b. Hepatol. Res. 2016, 46, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Karino, Y.; Toyota, J.; Ikeda, K.; Suzuki, F.; Chayama, K.; Kawakami, Y.; Ishikawa, H.; Watanabe, H.; Hernandez, D.; Yu, F.; et al. Characterization of virologic escape in hepatitis C virus genotype 1b patients treated with the direct-acting antivirals daclatasvir and asunaprevir. J. Hepatol. 2013, 58, 646–654. [Google Scholar] [CrossRef] [PubMed]
- Lenz, O.; Verbinnen, T.; Fevery, B.; Tambuyzer, L.; Vijgen, L.; Peeters, M.; Buelens, A.; Ceulemans, H.; Beumont, M.; Picchio, G.; et al. Virology analyses of HCV isolates from genotype 1-infected patients treated with simeprevir plus peginterferon/ribavirin in phase IIb/III studies. J. Hepatol. 2015, 62, 1008–1014. [Google Scholar] [CrossRef] [PubMed]
- McPhee, F.; Hernandez, D.; Yu, F.; Ueland, J.; Monikowski, A.; Carifa, A.; Falk, P.; Wang, C.; Fridell, R.; Eley, T.; et al. Resistance analysis of hepatitis C virus genotype 1 prior treatment null responders receiving daclatasvir and asunaprevir. Hepatology 2013, 58, 902–911. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.C.; de Meyer, S.; Bartels, D.J.; Dierynck, I.; Zhang, E.Z.; Spanks, J.; Tigges, A.M.; Ghys, A.; Dorrian, J.; Adda, N.; et al. Evolution of treatment-emergent resistant variants in telaprevir phase 3 clinical trials. Clin. Infect. Dis. 2013, 57, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Pawlotsky, J.M. Hepatitis C virus resistance to direct-acting antiviral drugs in interferon-free regimens. Gastroenterology 2016, 151, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Kumada, H.; Suzuki, Y.; Ikeda, K.; Toyota, J.; Karino, Y.; Chayama, K.; Kawakami, Y.; Ido, A.; Yamamoto, K.; Takaguchi, K.; et al. Daclatasvir plus asunaprevir for chronic HCV genotype 1b infection. Hepatology 2014, 59, 2083–2091. [Google Scholar] [CrossRef] [PubMed]
- Lontok, E.; Harrington, P.; Howe, A.; Kieffer, T.; Lennerstrand, J.; Lenz, O.; McPhee, F.; Mo, H.; Parkin, N.; Pilot-Matias, T.; et al. Hepatitis C virus drug resistance-associated substitutions: State of the art summary. Hepatology 2015, 62, 1623–1632. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.A.; Worth, A.; Martin, R.; Svarovskaia, E.; Brainard, D.M.; Lawitz, E.; Miller, M.D.; Mo, H. Characterization of hepatitis C virus resistance from a multiple-dose clinical trial of the novel NS5A inhibitor GS-5885. Antimicrob. Agents Chemother. 2013, 57, 6333–6340. [Google Scholar] [CrossRef] [PubMed]
- Zeuzem, S.; Mizokami, M.; Pianko, S.; Mangia, A.; Han, K.; Martin, R.; Svarovskaia, E.S.; Dvory-Sobol, H.; Doehle, B.; Pang, P.S.; et al. Prevalence of pre-treatment NS5A resistance associated variants in genotype 1 patients across different regions using deep sequencing and effect on treatment outcome with LDV/SOF. Hepatology 2015, 62, 254. [Google Scholar]
- Uchida, Y.; Kouyama, J.I.; Naiki, K.; Sugawarav, K.; Inao, M.; Imai, Y.; Nakayama, N.; Mochida, S. Development of rare RAVs that are extremely tolerant against NS5A inhibitors during daclatasvir/asunaprevir therapy via a two-hit mechanism. Hepatol. Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Yoshimi, S.; Imamura, M.; Murakami, E.; Hiraga, N.; Tsuge, M.; Kawakami, Y.; Aikata, H.; Abe, H.; Hayes, C.N.; Sasaki, T.; et al. Long term persistence of NS5A inhibitor-resistant hepatitis C virus in patients who failed daclatasvir and asunaprevir therapy. J. Med. Virol. 2015, 87, 1913–1920. [Google Scholar] [CrossRef] [PubMed]
- Sterling, R.K.; Lissen, E.; Clumeck, N.; Sola, R.; Correa, M.C.; Montaner, J.; M, S.S.; Torriani, F.J.; Dieterich, D.T.; Thomas, D.L.; et al. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology 2006, 43, 1317–1325. [Google Scholar] [CrossRef] [PubMed]
- Tadokoro, K.; Kobayashi, M.; Suzuki, F.; Tanaka, C.; Yamaguchi, T.; Nagano, M.; Egashira, T.; Kumada, H. Comparative quantitative analysis of hepatitis C mutations at amino acids 70 and 91 in the core region by the Q-invader assay. J. Virol. Methods 2013, 189, 221–227. [Google Scholar] [CrossRef] [PubMed]


| Parameter | Returned to HCV at Baseline | Continuous Predominant Substitution | p-Value |
|---|---|---|---|
| (n = 9) | (n = 7) | ||
| Age (years) a | 68 (51–74) | 65 (52–69) | 0.18 |
| Male/female | 4/5 | 3/4 | 1.00 |
| IL28B (rs8099917) TT/TG or GG | 1/8 | 3/4 | 0.26 |
| Hemoglobin (g/dL) a | 13.5 (12.0–15.3) | 13.6 (12.3–16.6) | 0.49 |
| Platelets (×104/μL) a | 16.1 (12.6–23.6) | 11.9 (8.3–17.5) | 0.03 |
| ALT (IU/L) a | 30 (17–73) | 60 (16–161) | 0.27 |
| γ-GT (IU/L) a | 24 (15–81) | 43 (17–96) | 0.34 |
| HCV-RNA (log IU/mL) a | 6.4 (5.6–7.4) | 6.7 (5.9–7.3) | 0.67 |
| Elastography (kPa) | 8.7 (3.1–10.0) | 6.8 (5.6–12.1) | 0.74 |
| FIB-4 index b | 2.7 (2.1–4.0) | 2.8 (2.0–4.9) | 0.96 |
| Response to SMV/PEG-IFN/RBV treatment (relapse/breakthrough) | 8/1 | 4/3 | 0.26 |
| Duration of follow up after treatment (week) a | 64 (33–78) | 66 (36–72) | 0.56 |
| No. | Background Liver Status | Previous Treatment | Response to DCV + ASV | Duration of DCV + ASV (Weeks) | RASs after DCV/ASV Treatment | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NS3/4A | NS5A | NS5B | |||||||||||
| Q80 | D168 | V170 | R30 | L31 | A92 | Y93 | Deletion | S282 | |||||
| AD-149 | CH | none | Relapse | 12 | Q | D | V | H | L | A | H | S | |
| AD-24 | Cirrhosis | none | Relapse | 24 | Q | D | I | R | V | A | H | S | |
| DCVF-18 | CH | PR | Relapse | 2 | Q | E/D | V | R | V | A | Y | S | |
| AD-58 | Cirrhosis | PR | Relapse | 5 | Q | D/E | M | R | L | A | Y | S | |
| DCVF-3 | CH | PR | Relapse | 24 | Q | Y | V | R | M | A | H | S | |
| AD-130 | CH | PR | Relapse | 2 | L | D | I | R | L | A | H | S | |
| DCVF-4 | Cirrhosis | PR | Relapse | 24 | Q | V | I | Q | L | A | H | S | |
| DCVF-7 | CH | none | BT | 20 | Q | D | V | R | I | A | H | S | |
| DCVF-16 | CH | none | BT | 14 | Q | D | V | Q | M | A | H | S | |
| DCVF-17 | CH | none | BT | 12 | Q | E | I | H | F | A | H | S | |
| AD-20 | CH | P | BT | 23 | Q | V | V | R | M | A | H | S | |
| DCVF-12 | Cirrhosis | PR | BT | 14 | Q | E | V | R | M | A | H | S | |
| AD-96 | Cirrhosis | PR | BT | 19 | Q | E | I | R | F | V | H | S | |
| DCVF-1 | CH | PR | BT | 14 | Q | V | I | Q | M | A | H | S | |
| DCVF-5 | CH | PR | BT | 11 | R | E | I | R/Q | M | A | H | S | |
| DCVF-11 | Cirrhosis | SMV + PR | BT | 10 | Q | E | V | R | V | A | H | S | |
| DCVF-15 | Cirrhosis | SMV + PR | BT | 10 | Q | V | V | R | F | A | N | S | |
| DCVF-9 | Cirrhosis | SMV + PR | BT | 12 | Q | V | V | R | M | A | H | S | |
| DCVF-8 | CH | SMV + PR | BT | 14 | Q | V | V | R | V | A | H | S | |
| DCVF-14 | Cirrhosis | SMV + PR | BT | 14 | L/R | E | I | R | V | A | H | S | |
| AD-25 | CH | SMV + PR | BT | 22 | Q/R | E | I | R | V | A | H | S | |
| DCVF-13 | Cirrhosis | SMV + PR | BT | 8 | K | D | I | Q | L/M | K/E/T/A | Y/H | S | |
| DCVF-6 | CH | SMV + PR | BT | 4 | Q | V | V | R | L | A | Y | Delete 29 | S |
| DCVF-2 | CH | SMV + PR | BT | 12 | K | E | I | R | L | A | Y | Delete 32 | S |
| DCVF-10 | Cirrhosis | SMV + PR | BT | 14 | Q | V | I | R | L/V | A | Y | Delete 32 | S |
| Parameter | Reverted to HCV at Baseline | Continuous Predominant Substitution | p-Value |
|---|---|---|---|
| (n = 6) | (n = 9) | ||
| Age (years) a | 68 (41–71) | 70 (49–76) | 0.29 |
| Male/female | 2/4 | 6/3 | 0.31 |
| IL28B (rs8099917) TT/TG or GG | 2/4 | 4/5 | 1.00 |
| Hemoglobin (g/dL) a | 13.1 (11.5–14.8) | 13.3 (11.9–15.5) | 0.72 |
| Platelets (× 104/µL) a | 16.1 (12.5–26.0) | 9.3 (6.6–21.8) | 0.03 |
| Prothrombin time (%) a | 99 (94–106) | 84 (60–97) | 0.03 |
| Total bilirubin (IU/L) a | 0.5 (0.3–0.5) | 0.7 (0.4–1.5) | 0.04 |
| ALT (IU/L) a | 46 (11–67) | 35 (13–85) | 0.91 |
| γ-GT (IU/L) a | 35 (14–57) | 32 (12–96) | 0.72 |
| Alb (mg/mL) a | 4.2 (3.9-4.4) | 3.9 (2.8-4.6) | 0.04 |
| HCV-RNA (log IU/mL) a | 6.4 (5.9–7.0) | 6.1 (5.6–6.8) | 0.19 |
| FIB-4 index b | 2.6 (1.1-4.2) | 4.3 (1.3-10.0) | 0.04 |
| Response to DCV/ASV treatment (relapse/breakthrough) | 2/4 | 1/8 | 0.53 |
| Follow-up duration after DCV/ASV treatment (week) a | 72 (46–231) | 85 (41–90) | 0.64 |
| Parameter | SMV/PEG-IFN/RBV Failure | DCV/ASV Failure |
|---|---|---|
| n = 20 | n = 25 | |
| Age (years) a | 66 (40–74) | 68 (41–78) |
| Male/female | 10/10 | 16/9 |
| Previous treatment | ||
| None | 6 (30%) | 5 (20%) |
| Conventional IFN | 14 (70%) | 10 (40%) |
| SMV/PEG-IFN/RBV | 0 (0%) | 10 (40%) |
| IL-28B (rs8099917) | ||
| TT | 5 (25%) | 11 (44%) |
| TG or GG | 15 (75%) | 14 (56%) |
| Hemoglobin (g/dL) a | 13.8 (12.0–16.6) | 13.1 (10.0–15.5) |
| Platelets (×104/μL) a | 15.5 (8.3–25.4) | 13.2 (3.9–31.5) |
| ALT (IU/L) a | 36.5 (16–161) | 35 (10–85) |
| γ-GT (IU/L) a | 35 (15–96) | 38 (12–96) |
| HCV-RNA (Log IU/mL) a | 6.8 (5.6–7.5) | 6.1 (4.6–7.0) |
| Background liver status | ||
| Chronic hepatitis | 20 (100%) | 11 (44%) |
| Cirrhosis | 0 (0%) | 14 (56%) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshida, K.; Hai, H.; Tamori, A.; Teranishi, Y.; Kozuka, R.; Motoyama, H.; Kawamura, E.; Hagihara, A.; Uchida-Kobayashi, S.; Morikawa, H.; et al. Long-Term Follow-Up of Resistance-Associated Substitutions in Hepatitis C Virus in Patients in Which Direct Acting Antiviral-Based Therapy Failed. Int. J. Mol. Sci. 2017, 18, 962. https://doi.org/10.3390/ijms18050962
Yoshida K, Hai H, Tamori A, Teranishi Y, Kozuka R, Motoyama H, Kawamura E, Hagihara A, Uchida-Kobayashi S, Morikawa H, et al. Long-Term Follow-Up of Resistance-Associated Substitutions in Hepatitis C Virus in Patients in Which Direct Acting Antiviral-Based Therapy Failed. International Journal of Molecular Sciences. 2017; 18(5):962. https://doi.org/10.3390/ijms18050962
Chicago/Turabian StyleYoshida, Kanako, Hoang Hai, Akihiro Tamori, Yuga Teranishi, Ritsuzo Kozuka, Hiroyuki Motoyama, Etsushi Kawamura, Atsushi Hagihara, Sawako Uchida-Kobayashi, Hiroyasu Morikawa, and et al. 2017. "Long-Term Follow-Up of Resistance-Associated Substitutions in Hepatitis C Virus in Patients in Which Direct Acting Antiviral-Based Therapy Failed" International Journal of Molecular Sciences 18, no. 5: 962. https://doi.org/10.3390/ijms18050962






