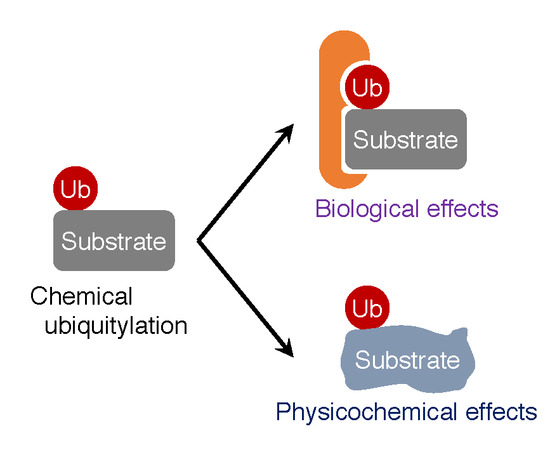
Biological and Physicochemical Functions of Ubiquitylation Revealed by Synthetic Chemistry Approaches
Abstract
:1. Introduction
2. Strategies for Chemical Preparation of Ubiquitylated Proteins
2.1. Native Chemical Ligation and Isopeptide Chemical Ligation
2.2. Ubiquitin Conjugation via Non-Native Linkages
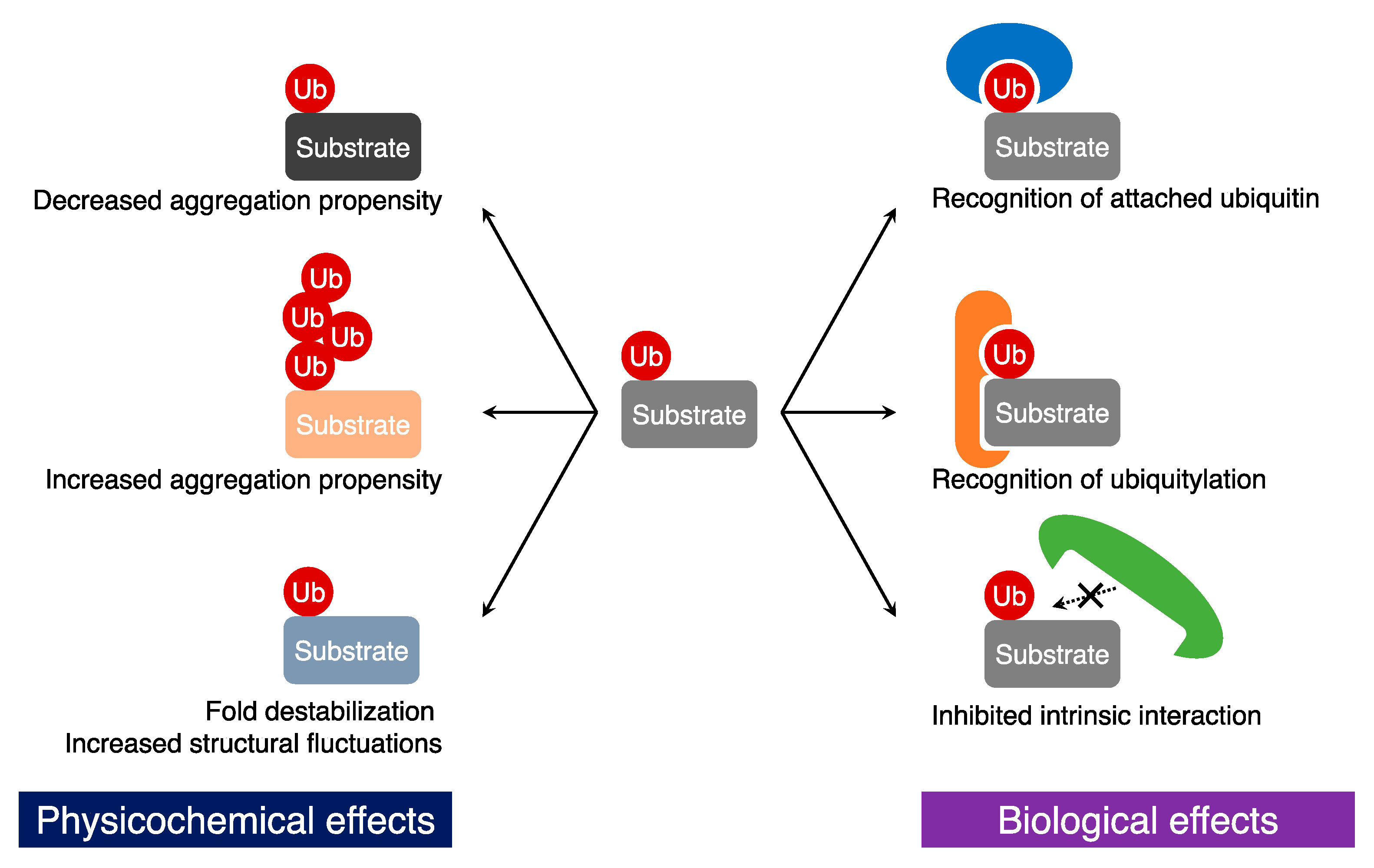
3. Biological Effects of Ubiquitylation on Substrates
3.1. Recognition of Ubiquitin Tags
3.2. Generation of New Protein-Protein Interactions
3.3. Inhibition of Protein-Protein Interactions
4. Physicochemical Effects of Ubiquitylation on Target Proteins
4.1. Changes in Aggregation Propensity
4.2. Fold Destabilization and Structural Fluctuation
5. Conclusions
Acknowledgments
Conflicts of Interest
Abbreviations
| DUBs | Deubiquitinating enzymes |
| E1 | Ubiquitin-activating enzymes |
| E2 | Ubiquitin-conjugating enzymes |
| E3 | Ubiquitin ligases |
| NCL | Native chemical ligation |
| UFD | Ubiquitin fusion degradation |
References
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.H.; Yeh, L.S.; Huang, H.; Arminski, L.; Castro-Alvear, J.; Chen, Y.; Hu, Z.; Kourtesis, P.; Ledley, R.S.; Suzek, B.E.; et al. The protein information resource. Nucleic Acids Res. 2003, 31, 345–347. [Google Scholar] [CrossRef] [PubMed]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. Phosphositeplus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.S.; Ma, P.C.M.; Ota, I.M.; Varshavsky, A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 1995, 270, 17442–17456. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Ben-Saadon, R. N-terminal ubiquitination: More protein substrates join in. Trends Cell Biol. 2004, 14, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Schulman, B.A.; Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009, 10, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Joazeiro, C.A. Ring domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef] [PubMed]
- Keren-Kaplan, T.; Attali, I.; Motamedchaboki, K.; Davis, B.A.; Tanner, N.; Reshef, Y.; Laudon, E.; Kolot, M.; Levin-Kravets, O.; Kleifeld, O.; et al. Synthetic biology approach to reconstituting the ubiquitylation cascade in bacteria. EMBO J. 2012, 31, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, D.; Isogai, S.; Tenno, T.; Tochio, H.; Shirakawa, M.; Ariyoshi, M. Purification, crystallization and preliminary crystallographic studies of LYS48-linked polyubiquitin chains. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 834–837. [Google Scholar] [CrossRef] [PubMed]
- Spasser, L.; Brik, A. Chemistry and biology of the ubiquitin signal. Angew. Chem. Int. Ed. Engl. 2012, 51, 6840–6862. [Google Scholar] [CrossRef] [PubMed]
- Strieter, E.R.; Korasick, D.A. Unraveling the complexity of ubiquitin signaling. ACS Chem. Biol. 2012, 7, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Hemantha, H.P.; Brik, A. Non-enzymatic synthesis of ubiquitin chains: Where chemistry makes a difference. Bioorg. Med. Chem. 2013, 21, 3411–3420. [Google Scholar] [CrossRef] [PubMed]
- Abeywardana, T.; Pratt, M.R. Using chemistry to investigate the molecular consequences of protein ubiquitylation. ChemBioChem 2014, 15, 1547–1554. [Google Scholar] [CrossRef] [PubMed]
- Faggiano, S.; Pastore, A. The challenge of producing ubiquitinated proteins for structural studies. Cells 2014, 3, 639–656. [Google Scholar] [CrossRef] [PubMed]
- Weller, C.E.; Pilkerton, M.E.; Chatterjee, C. Chemical strategies to understand the language of ubiquitin signaling. Biopolymers 2014, 101, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Pham, G.H.; Strieter, E.R. Peeling away the layers of ubiquitin signaling complexities with synthetic ubiquitin-protein conjugates. Curr. Opin. Chem. Biol. 2015, 28, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Liu, C.F. Chemical methods for protein ubiquitination. Top. Curr. Chem. 2015, 362, 89–106. [Google Scholar] [PubMed]
- Valkevich, E.M.; Guenette, R.G.; Sanchez, N.A.; Chen, Y.C.; Ge, Y.; Strieter, E.R. Forging isopeptide bonds using thiol-ene chemistry: Site-specific coupling of ubiquitin molecules for studying the activity of isopeptidases. J. Am. Chem. Soc. 2012, 134, 6916–6919. [Google Scholar] [CrossRef] [PubMed]
- Trang, V.H.; Valkevich, E.M.; Minami, S.; Chen, Y.C.; Ge, Y.; Strieter, E.R. Nonenzymatic polymerization of ubiquitin: Single-step synthesis and isolation of discrete ubiquitin oligomers. Angew. Chem. Int. Ed. Engl. 2012, 51, 13085–13088. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Krantz, B.; Russell, N.S.; Deshpande, S.; Wilkinson, K.D. Nonhydrolyzable diubiquitin analogues are inhibitors of ubiquitin conjugation and deconjugation. Biochemistry 2000, 39, 10001–10010. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.; Schneider, D.; Rosner, D.; Malhotra, S.; Mortensen, F.; Mayer, T.U.; Scheffner, M.; Marx, A. Dissecting ubiquitin signaling with linkage-defined and protease resistant ubiquitin chains. Angew. Chem. Int. Ed. Engl. 2014, 53, 12925–12929. [Google Scholar] [CrossRef] [PubMed]
- Eger, S.; Castrec, B.; Hubscher, U.; Scheffner, M.; Rubini, M.; Marx, A. Generation of a mono-ubiquitinated PCNA mimic by click chemistry. ChemBioChem 2011, 12, 2807–2812. [Google Scholar] [CrossRef] [PubMed]
- Eger, S.; Scheffner, M.; Marx, A.; Rubini, M. Synthesis of defined ubiquitin dimers. J. Am. Chem. Soc. 2010, 132, 16337–16339. [Google Scholar] [CrossRef] [PubMed]
- Weikart, N.D.; Sommer, S.; Mootz, H.D. Click synthesis of ubiquitin dimer analogs to interrogate linkage-specific UBA domain binding. Chem. Commun. (Camb.) 2012, 48, 296–298. [Google Scholar] [CrossRef] [PubMed]
- Shanmugham, A.; Fish, A.; Luna-Vargas, M.P.; Faesen, A.C.; El Oualid, F.; Sixma, T.K.; Ovaa, H. Nonhydrolyzable ubiquitin-isopeptide isosteres as deubiquitinating enzyme probes. J. Am. Chem. Soc. 2010, 132, 8834–8835. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, C.; McGinty, R.K.; Fierz, B.; Muir, T.W. Disulfide-directed histone ubiquitylation reveals plasticity in hdot1L activation. Nat. Chem. Biol. 2010, 6, 267–269. [Google Scholar] [CrossRef] [PubMed]
- Fierz, B.; Chatterjee, C.; McGinty, R.K.; Bar-Dagan, M.; Raleigh, D.P.; Muir, T.W. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 2011, 7, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ai, Y.; Wang, J.; Haracska, L.; Zhuang, Z. Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nat. Chem. Biol. 2010, 6, 270–272. [Google Scholar] [CrossRef] [PubMed]
- Meier, F.; Abeywardana, T.; Dhall, A.; Marotta, N.P.; Varkey, J.; Langen, R.; Chatterjee, C.; Pratt, M.R. Semisynthetic, site-specific ubiquitin modification of α-synuclein reveals differential effects on aggregation. J. Am. Chem. Soc. 2012, 134, 5468–5471. [Google Scholar] [CrossRef] [PubMed]
- Abeywardana, T.; Lin, Y.H.; Rott, R.; Engelender, S.; Pratt, M.R. Site-specific differences in proteasome-dependent degradation of monoubiquitinated α-synuclein. Chem. Biol. 2013, 20, 1207–1213. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.; Lewis, S.M.; Sasaki, A.T.; Wilkerson, E.M.; Locasale, J.W.; Cantley, L.C.; Kuhlman, B.; Dohlman, H.G.; Campbell, S.L. Site-specific monoubiquitination activates RAS by impeding GTPase-activating protein function. Nat. Struct. Mol. Biol. 2013, 20, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, D.; Walinda, E.; Fukada, H.; Sugase, K.; Shirakawa, M. Ubiquitylation directly induces fold destabilization of proteins. Sci. Rep. 2016, 6, 39453. [Google Scholar] [CrossRef] [PubMed]
- Hemantha, H.P.; Bavikar, S.N.; Herman-Bachinsky, Y.; Haj-Yahya, N.; Bondalapati, S.; Ciechanover, A.; Brik, A. Nonenzymatic polyubiquitination of expressed proteins. J. Am. Chem. Soc. 2014, 136, 2665–2673. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.E.; Chudasama, V.; Moody, P.; Smith, M.E.; Caddick, S. A novel synthetic chemistry approach to linkage-specific ubiquitin conjugation. Org. Biomol. Chem. 2015, 13, 4165–4168. [Google Scholar] [CrossRef] [PubMed]
- Ajish Kumar, K.S.; Haj-Yahya, M.; Olschewski, D.; Lashuel, H.A.; Brik, A. Highly efficient and chemoselective peptide ubiquitylation. Angew. Chem. Int. Ed. Engl. 2009, 48, 8090–8094. [Google Scholar] [CrossRef] [PubMed]
- Erlich, L.A.; Kumar, K.S.; Haj-Yahya, M.; Dawson, P.E.; Brik, A. N-methylcysteine-mediated total chemical synthesis of ubiquitin thioester. Org. Biomol. Chem. 2010, 8, 2392–2396. [Google Scholar] [CrossRef] [PubMed]
- Hejjaoui, M.; Haj-Yahya, M.; Kumar, K.S.; Brik, A.; Lashuel, H.A. Towards elucidation of the role of ubiquitination in the pathogenesis of Parkinson’s disease with semisynthetic ubiquitinated α-synuclein. Angew. Chem. Int. Ed. Engl. 2011, 50, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Haj-Yahya, M.; Fauvet, B.; Herman-Bachinsky, Y.; Hejjaoui, M.; Bavikar, S.N.; Karthikeyan, S.V.; Ciechanover, A.; Lashuel, H.A.; Brik, A. Synthetic polyubiquitinated α-synuclein reveals important insights into the roles of the ubiquitin chain in regulating its pathophysiology. Proc. Natl. Acad. Sci. USA 2013, 110, 17726–17731. [Google Scholar] [CrossRef] [PubMed]
- Shabek, N.; Herman-Bachinsky, Y.; Buchsbaum, S.; Lewinson, O.; Haj-Yahya, M.; Hejjaoui, M.; Lashuel, H.A.; Sommer, T.; Brik, A.; Ciechanover, A. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol. Cell 2012, 48, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Fekner, T.; Ottesen, J.J.; Chan, M.K. A pyrrolysine analogue for site-specific protein ubiquitination. Angew. Chem. Int. Ed. Engl. 2009, 48, 9184–9187. [Google Scholar] [CrossRef] [PubMed]
- Madrzak, J.; Fiedler, M.; Johnson, C.M.; Ewan, R.; Knebel, A.; Bienz, M.; Chin, J.W. Ubiquitination of the Dishevelled DIX domain blocks its head-to-tail polymerization. Nat. Commun. 2015, 6, 6718. [Google Scholar] [CrossRef] [PubMed]
- Fierz, B.; Kilic, S.; Hieb, A.R.; Luger, K.; Muir, T.W. Stability of nucleosomes containing homogenously ubiquitylated H2A and H2B prepared using semisynthesis. J. Am. Chem. Soc. 2012, 134, 19548–19551. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, C.; McGinty, R.K.; Pellois, J.P.; Muir, T.W. Auxiliary-mediated site-specific peptide ubiquitylation. Angew. Chem. Int. Ed. Engl. 2007, 46, 2814–2818. [Google Scholar] [CrossRef] [PubMed]
- McGinty, R.K.; Kim, J.; Chatterjee, C.; Roeder, R.G.; Muir, T.W. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 2008, 453, 812–816. [Google Scholar] [CrossRef] [PubMed]
- McGinty, R.K.; Kohn, M.; Chatterjee, C.; Chiang, K.P.; Pratt, M.R.; Muir, T.W. Structure-activity analysis of semisynthetic nucleosomes: Mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B. ACS Chem. Biol. 2009, 4, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Pasunooti, K.K.; Li, F.; Liu, X.W.; Liu, C.F. Dual native chemical ligation at Lysine. J. Am. Chem. Soc. 2009, 131, 13592–13593. [Google Scholar] [CrossRef] [PubMed]
- Weller, C.E.; Huang, W.; Chatterjee, C. Facile synthesis of native and protease-resistant ubiquitylated peptides. ChemBioChem 2014, 15, 1263–1267. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Bi, X.; Li, F.; Cao, Y.; Liu, C.F. Native chemical ubiquitination using a genetically incorporated azidonorleucine. Chem. Commun. (Camb.) 2014, 50, 7971–7974. [Google Scholar] [CrossRef] [PubMed]
- Castaneda, C.; Liu, J.; Chaturvedi, A.; Nowicka, U.; Cropp, T.A.; Fushman, D. Nonenzymatic assembly of natural polyubiquitin chains of any linkage composition and isotopic labeling scheme. J. Am. Chem. Soc. 2011, 133, 17855–17868. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Pasunooti, K.K.; Li, F.; Liu, X.W.; Liu, C.F. Synthesis of K48-linked diubiquitin using dual native chemical ligation at lysine. Chem. Commun. (Camb.) 2010, 46, 7199–7201. [Google Scholar] [CrossRef] [PubMed]
- Merkx, R.; de Bruin, G.; Kruithof, A.; van den Bergh, T.; Snip, E.; Lutz, M.; El Oualid, F.; Ovaa, H. Scalable synthesis of γ-thiolysine starting from lysine and a side by side comparison with δ-thiolysine in non-enzymatic ubiquitination. Chem. Sci. 2013, 4, 4494. [Google Scholar] [CrossRef]
- Kumar, K.S.; Spasser, L.; Erlich, L.A.; Bavikar, S.N.; Brik, A. Total chemical synthesis of di-ubiquitin chains. Angew. Chem. Int. Ed. Engl. 2010, 49, 9126–9131. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.S.; Bavikar, S.N.; Spasser, L.; Moyal, T.; Ohayon, S.; Brik, A. Total chemical synthesis of a 304 amino acid K48-linked tetraubiquitin protein. Angew. Chem. Int. Ed. Engl. 2011, 50, 6137–6141. [Google Scholar] [CrossRef] [PubMed]
- Bavikar, S.N.; Spasser, L.; Haj-Yahya, M.; Karthikeyan, S.V.; Moyal, T.; Kumar, K.S.; Brik, A. Chemical synthesis of ubiquitinated peptides with varying lengths and types of ubiquitin chains to explore the activity of deubiquitinases. Angew. Chem. Int. Ed. Engl. 2012, 51, 758–763. [Google Scholar] [CrossRef] [PubMed]
- Virdee, S.; Kapadnis, P.B.; Elliott, T.; Lang, K.; Madrzak, J.; Nguyen, D.P.; Riechmann, L.; Chin, J.W. Traceless and site-specific ubiquitination of recombinant proteins. J. Am. Chem. Soc. 2011, 133, 10708–10711. [Google Scholar] [CrossRef] [PubMed]
- Virdee, S.; Ye, Y.; Nguyen, D.P.; Komander, D.; Chin, J.W. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat. Chem. Biol. 2010, 6, 750–757. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, T.; Mishima, Y.; Hojo, H.; Suetake, I. Synthesis of ubiquitylated histone h3 using a thiirane linker for chemical ligation. J. Pept. Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; He, Q.; Liu, Y.; Liu, S.; Tang, S.; Li, C.; Sun, D.; Li, X.; Zhou, M.; Zhu, P.; et al. Chemical synthesis of K34-ubiquitylated H2B for nucleosome reconstitution and single-particle cryo-electron microscopy structural analysis. ChemBioChem 2017, 18, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Gao, S.; Zheng, Y.; Tan, X.; Lan, H.; Tan, X.; Sun, D.; Lu, L.; Wang, T.; Zheng, Q.; et al. Quasi-racemic X-ray structures of K27-linked ubiquitin chains prepared by total chemical synthesis. J. Am. Chem. Soc. 2016, 138, 7429–7435. [Google Scholar] [CrossRef] [PubMed]
- Dawson, P.E.; Muir, T.W.; Clark-Lewis, I.; Kent, S.B. Synthesis of proteins by native chemical ligation. Science 1994, 266, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Husnjak, K.; Dikic, I. Ubiquitin-binding proteins: Decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 2012, 81, 291–322. [Google Scholar] [CrossRef] [PubMed]
- Chau, V.; Tobias, J.W.; Bachmair, A.; Marriott, D.; Ecker, D.J.; Gonda, D.K.; Varshavsky, A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 1989, 243, 1576–1583. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Williamson, A.; Banerjee, S.; Philipp, I.; Rape, M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 2008, 133, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Shabek, N.; Iwai, K.; Ciechanover, A. Ubiquitin is degraded by the ubiquitin system as a monomer and as part of its conjugated target. Biochem. Biophys. Res. Commun. 2007, 363, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Kannouche, P.L.; Wing, J.; Lehmann, A.R. Interaction of human DNA polymerase η with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 2004, 14, 491–500. [Google Scholar] [CrossRef]
- Watanabe, K.; Tateishi, S.; Kawasuji, M.; Tsurimoto, T.; Inoue, H.; Yamaizumi, M. Rad18 guides polη to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004, 23, 3886–3896. [Google Scholar] [CrossRef] [PubMed]
- Schwarz-Romond, T.; Fiedler, M.; Shibata, N.; Butler, P.J.; Kikuchi, A.; Higuchi, Y.; Bienz, M. The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat. Struct. Mol. Biol. 2007, 14, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.P.; Walker, D.E.; Goldstein, J.M.; de Laat, R.; Banducci, K.; Caccavello, R.J.; Barbour, R.; Huang, J.; Kling, K.; Lee, M.; et al. Phosphorylation of Ser-129 is the dominant pathological modification of α-synuclein in familial and sporadic Lewy body disease. J. Biol. Chem. 2006, 281, 29739–29752. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Fujiwara, H.; Nonaka, T.; Wakabayashi, K.; Takahashi, H.; Lee, V.M.; Trojanowski, J.Q.; Mann, D.; Iwatsubo, T. Phosphorylated α-synuclein is ubiquitinated in α-synucleinopathy lesions. J. Biol. Chem. 2002, 277, 49071–49076. [Google Scholar] [CrossRef] [PubMed]
- Hagai, T.; Levy, Y. Ubiquitin not only serves as a tag but also assists degradation by inducing protein unfolding. Proc. Natl. Acad. Sci. USA 2010, 107, 2001–2006. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.J.; Rape, M. Enhanced protein degradation by branched ubiquitin chains. Cell 2014, 157, 910–921. [Google Scholar] [CrossRef] [PubMed]
- Ohtake, F.; Saeki, Y.; Ishido, S.; Kanno, J.; Tanaka, K. The K48-K63 branched ubiquitin chain regulates NF-κB signaling. Mol. Cell 2016, 64, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liu, W.; Ye, Y.; Li, W. Ufd2p synthesizes branched ubiquitin chains to promote the degradation of substrates modified with atypical chains. Nat. Commun. 2017, 8, 14274. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Hod, Y.; Hershko, A. Heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem. Biophys. Res. Commun. 1978, 81, 1100–1105. [Google Scholar] [CrossRef]

| Method | Donor Ub | Acceptor | Difference | Reference |
|---|---|---|---|---|
| Co-expression | Ub wt | Mind bomb | – | Keren-Kaplan et al. 2012 [8] |
| Fusion proteins | Ub G76V | β-gal | N-terminal ubiquitylation | Johnson et al. 1995 [4] |
| Alkene-thiol reaction/chloroketone-thiol | Ub alkene | Ub Cys | Thioether linkage | Valkevich et al. 2012 [18] |
| Ub Cys allylamine | Ub Cys allylamine | Trang et al. 2012 [19] | ||
| Ub G76C | Ub Cys | Yin et al. 2000 [20] | ||
| Alkyne-azide reaction | Ub azide | Polyβ Plk | Triazole linkage | Schneider et al. 2014 [21] |
| PCNA Plk | Eger et al. 2011 [22] | |||
| Ub Plk | Eger et al. 2010 [23] | |||
| Ub alkyne | Ub AzF | Weikart et al. 2012 [24] | ||
| Aminoxy-aldehyde reaction | Ub aldehyde | Ub aminoxy | Oxime linkage | Shanmugham et al. 2010 [25] |
| Disulfide conjugation | Ub C-terminal aminoethanethiol linker | Histone H2B Cys | Disulfide bridge | Chatterjee et al. 2010 [26]; Fierz et al. 2011[27] |
| PCNA Cys | Chen et al. 2010 [28] | |||
| α-synuclein Cys | Meier et al. 2012 [29]; Abeywardana et al. 2013 [30] | |||
| Ub G76C | Ras Cys | Baker et al. 2013 [31] | ||
| CaM Cys, FABP4 Cys, FKBP12 Cys | Morimoto et al. 2016 [32] | |||
| Ubhydrazide | α-globin Cys | Hemantha et al. 2014 [33] | ||
| Maleimide-thiol reaction | Ub G76C | Ub Cys | Maleimide linkage Pyridazinedione linkage | Morgan et al. 2015 [34] |
| NCL/ICL | Ub-α-thioester | α-synuclein δ-thiolysine | – | Kumar et al. 2009 [35]; Erlich et al. 2010 [36]; Hejjaoui et al. 2011 [37]; Haj-Yahya et al. 2013 [38]; Shabek et al. 2012 [39] |
| CaM pyrrolysine analogue | Cysteine at residue 76 of ubiquitin | Li et al. 2009 [40] | ||
| Dvl2 DIX Nε−(t−butyloxycarbonyl)−l−lysine | – | Madrzak et al. 2015 [41] | ||
| Histone H2A Nε−(D−cysteinyl)−l−lysine | Alanine at residue 76 of ubiquitin | Fierz et al. 2012 [42] | ||
| Histone H2B thiol−auxiliary | – | Chatterjee et al. 2007[43]; McGinty et al. 2008[44] | ||
| Alanine at residue 76 of ubiquitin | McGinty et al. 2009 [45] | |||
| Peptide γ−thiolysine | – | Yang et al. 2009 [46] | ||
| Peptide thiol−auxiliary | Weller et al. 2014 [47] | |||
| Ub azidonorleucine | Yang et al. 2014 [48] | |||
| Ub Boc−lysine | Castaneda et al. 2011 [49] | |||
| Ub γ−thiolysine | Yang et al. 2010 [50] | |||
| Ub δ/γ−thiolysine | Merkx et al. 2013 [51] | |||
| Ub δ−thiolysine | Kumar et al. 2010 [52]; Kumar et al. 2011 [53]; Bavikar et al. 2012 [54]; Virdee et al. 2011 [55] | |||
| Ub Nε−(t−butyloxycarbonyl)−l−lysine | Virdee et al. 2010 [56] | |||
| Ub Cys-Pro-ester | Histone H3 Cys | Thioether linkage | Kawakami et al. 2017 [57] | |
| Ub hydrazide | Histone H2B thiol−auxiliary | – | Li et al. 2017 [58] | |
| Ub glycyl−auxiliary | Pan et al. 2016 [59] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morimoto, D.; Walinda, E.; Sugase, K.; Shirakawa, M. Biological and Physicochemical Functions of Ubiquitylation Revealed by Synthetic Chemistry Approaches. Int. J. Mol. Sci. 2017, 18, 1145. https://doi.org/10.3390/ijms18061145
Morimoto D, Walinda E, Sugase K, Shirakawa M. Biological and Physicochemical Functions of Ubiquitylation Revealed by Synthetic Chemistry Approaches. International Journal of Molecular Sciences. 2017; 18(6):1145. https://doi.org/10.3390/ijms18061145
Chicago/Turabian StyleMorimoto, Daichi, Erik Walinda, Kenji Sugase, and Masahiro Shirakawa. 2017. "Biological and Physicochemical Functions of Ubiquitylation Revealed by Synthetic Chemistry Approaches" International Journal of Molecular Sciences 18, no. 6: 1145. https://doi.org/10.3390/ijms18061145