De Novo Transcriptome Sequencing and the Hypothetical Cold Response Mode of Saussurea involucrata in Extreme Cold Environments
Abstract
:1. Introduction
2. Results and Discussion
2.1. Transcriptome Sequencing, Assembly, and Annotation
2.2. Differential Expression Analysis of the Assembled S. involucrata Transcripts under Different Chilling Temperatures
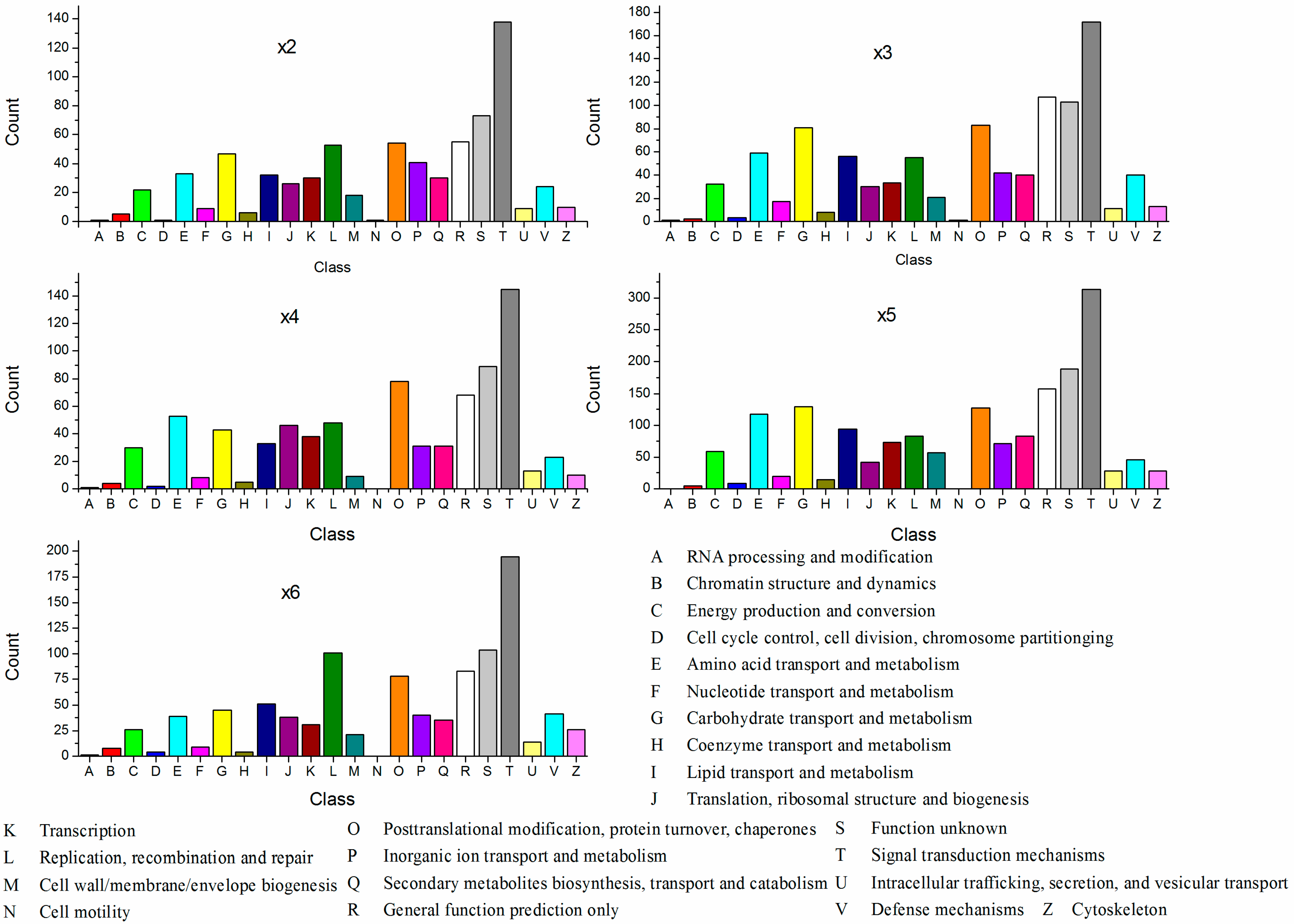
2.3. Cluster of Orthologous Groups of Proteins Classification of the Differentially Expressed Genes
2.4. The Transcriptional Regulatory Network Is Involved in the Adaptation of S. involucrata to Extreme Cold Environments
2.5. Cold Regulation of Signal Transduction Components
2.6. Membrane Proteins Associated with Transport Play Important Roles in Cold Adaptation
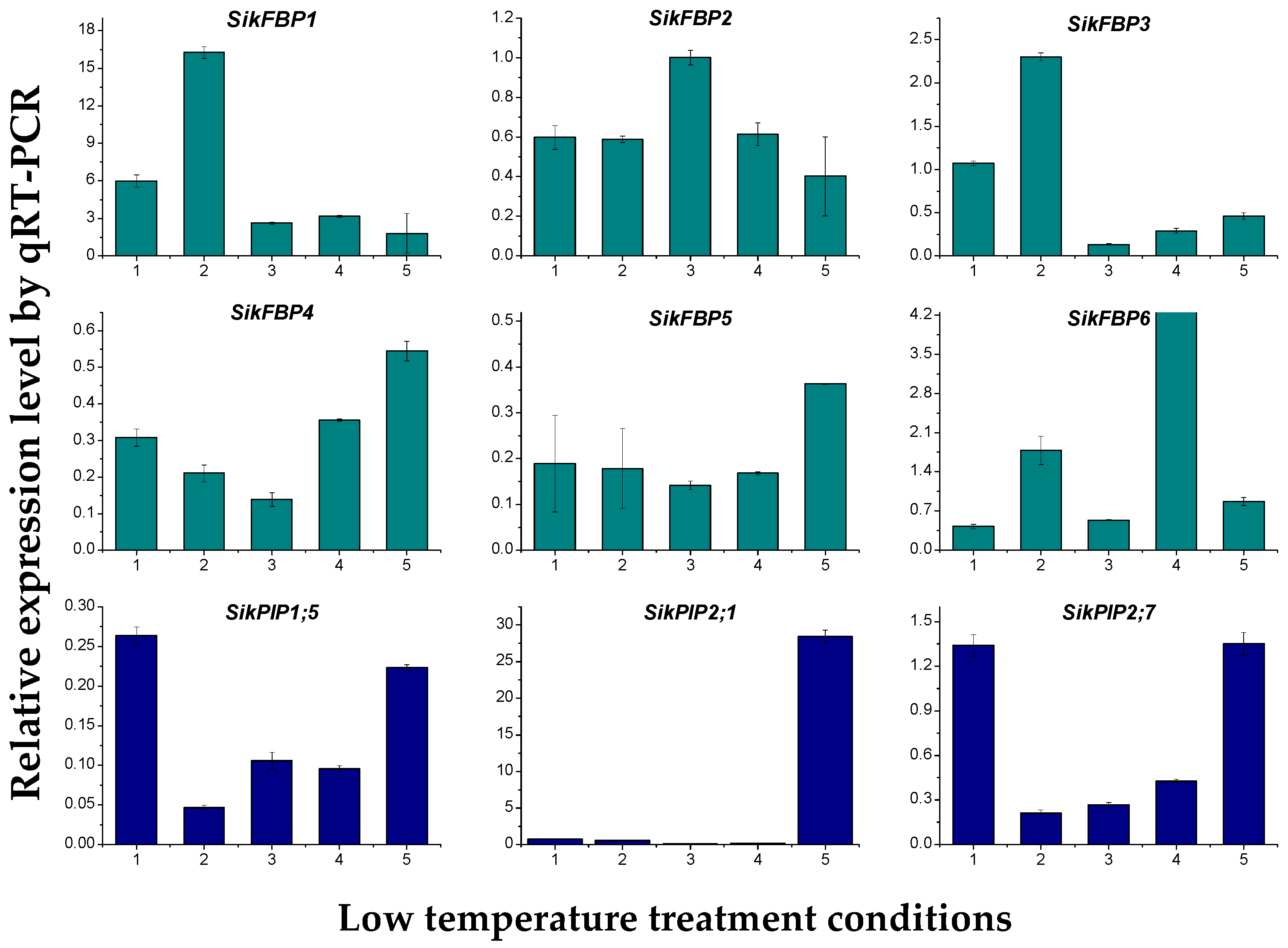
2.7. Quantitative Real-Time-PCR Validation of Differentially Expressed Transcripts from RNA-Seq
3. Materials and Methods
3.1. Plant Materials and Stress Treatment
3.2. Total RNA Extraction and Sequencing
3.3. Transcriptome Assembly and Annotation
3.4. Differential Expression Analysis of Transcripts
3.5. Validation of RNA-Seq Data by qRT-PCR
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| TF | Transcription factor |
| PE | Paired end |
| NGS | Next-generation sequencing |
| qRT-PCR | Quantitative real-time PCR |
| DEG | Differentially expressed gene |
| RPKM | Reads per kilo bases per million mapped reads |
References
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Naya, L.; Ladrera, R.; Ramos, J.; Gonzalez, E.M.; Arrese-Igor, C.; Minchin, F.R.; Becana, M. The response of carbon metabolism and antioxidant defenses of alfalfa nodules to drought stress and to the subsequent recovery of plants. Plant Physiol. 2007, 144, 1104–1114. [Google Scholar] [CrossRef]
- Veatch, M.E.; Smith, S.E.; Vandemark, G. Shoot biomass production among accessions of exposed to NaCl. Crop Sci. 2004, 44, 1008–1013. [Google Scholar] [CrossRef]
- Li, D.; Zhang, Y.; Hu, X.; Shen, X.; Ma, L.; Su, Z.; Wang, T.; Dong, J. Transcriptional profiling of Medicago truncatula under salt stress identified a novel CBF transcription factor MtCBF4 that plays an important role in abiotic stress responses. BMC Plant Biol. 2011, 11, 109. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Chen, Y.; Zhao, M.; Zhang, W.H. A novel Medicago truncatula HD-Zip gene, MtHB2, is involved in abiotic stress responses. Environ. Exp. Bot. 2012, 80, 1–9. [Google Scholar] [CrossRef]
- Guy, C.L. Cold acclimation and freezing stress tolerance: Role of protein metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1990, 41, 187–223. [Google Scholar] [CrossRef]
- Obata, T.; Fernie, A.R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef] [PubMed]
- Ingram, J.; Bartels, D. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 377–403. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta. 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.L.; Shen, H.T.; Chen, C.; Zhou, X.R.; Liu, H.F.; Zhu, J.B. Identification of a putative stearoyl acyl-carrier-protein desaturase gene from Saussurea involucrata Kar. et Kir. Biol. Plant. 2015, 59, 316–324. [Google Scholar] [CrossRef]
- Qiu, H.L.; Zhang, L.H.; Liu, C.; He, L.; Wang, A.Y.; Liu, H.L.; Zhu, J.B. Cloning and characterization of a novel dehydrin gene, SiDhn2, from Saussurea involucrata Kar. et Kir. Plant Mol. Biol. 2014, 84, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.C. Next-generation sequencing transforms today’s biology. Nat. Methods 2008, 5, 16–18. [Google Scholar] [CrossRef] [PubMed]
- Denoeud, F.; Aury, J.M.; Da, S.C.; Noel, B.; Rogier, O.; Delledonne, M.; Morgante, M.; Valle, G.; Wincker, P.; Scarpelli, C.; et al. Annotating genomes with massive-scale RNA sequencing. Genome Biol. 2008, 9, R175. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Leyva-Gonzalez, M.A.; Ibarra-Laclette, E.; Cruz-Ramirez, A.; Herrera-Estrella, L. Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS ONE 2012, 7, e48138. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.N.; Shi, D.Q.; Ruan, M.B.; Zhang, L.L.; Meng, Z.H.; Liu, J.; Yang, W.C. Transcriptome analysis reveals crosstalk of responsive genes to multiple abiotic stresses in cotton (Gossypium hirsutum L.). PLoS ONE 2013, 8, e80218. [Google Scholar] [CrossRef] [PubMed]
- Limpens, E.; Moling, S.; Hooiveld, G.; Pereira, P.A.; Bisseling, T.; Becker, J.D.; Küster, H. Cell- and tissue-specific transcriptome analyses of Medicago truncatula root nodules. PLoS ONE 2013, 8, e64377. [Google Scholar] [CrossRef] [PubMed]
- Boscari, A.; del, G.J.; Ferrarini, A.; Venturini, L.; Zaffini, A.L.; Delledonne, M.; Puppo, A. Expression dynamics of the Medicago truncatula transcriptome during the symbiotic interaction with Sinorhizobium meliloti: Which role for nitricoxide? Plant Physiol. 2013, 161, 425–439. [Google Scholar] [CrossRef] [PubMed]
- Cheung, F.; Haas, B.J.; Goldberg, S.M.; May, G.D.; Xiao, Y.; Town, C.D. Sequencing Medicago truncatula expressed sequenced tags using 454 Life Sciences technology. BMC Genom. 2006, 7, 272. [Google Scholar] [CrossRef] [PubMed]
- Cabeza, R.A.; Liese, R.; Lingner, A.; Von, S.I.; Neumann, J.; Salinas-Riester, G.; Pommerenke, C.; Dittert, K.; Schulze, J. RNA-seq transcriptome profiling reveals that Medicago truncatula nodules acclimate N2 fixation before emerging P deficiency reaches the nodules. J. Exp. Bot. 2014, 65, 6035–6048. [Google Scholar] [CrossRef] [PubMed]
- De Michele, R.; Formentin, E.; Todesco, M.; Toppo, S.; Carimi, F.; Zottini, M.; Barizza, E.; Ferrarini, A.; Delledonne, M.; Fontana, P.; et al. Transcriptome analysis of Medicago truncatula leaf senescence: Similarities and differences in metabolic and transcriptional regulations as compared with Arabidopsis, nodule senescence and nitric oxide signaling. New Phytol. 2009, 181, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.M.; Kong, X.D.; Chen, F.; Huang, J.X.; Lou, X.Y.; Zhao, J.Y. Transcriptome analysis of Brassica napus pod using RNA-Seq and identification of lipid-related candidate genes. BMC Genom. 2015, 16, 858. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.S.; Tu, Z.J.; Cheung, F.; Xu, W.W.; Lamb, J.F.; Jung, H.J.; Vance, C.P.; Gronwald, J.W. Using RNA-Seq for gene identification, polymorphism detection and transcript profiling in two alfalfa genotypes with divergent cell wall composition in stems. BMC Genom. 2011, 12, 199. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Patel, R.K.; Tyagi, A.K.; Jain, M. De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res. 2011, 18, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Mizrachi, E.; Hefer, C.A.; Ranik, M.; Joubert, F.; Myburg, A.A. De novo assembled expressed gene catalog of a fast-growing Eucalyptus tree produced by Illumina mRNA-Seq. BMC Genom. 2010, 11, 681. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Fang, B.; Chen, J.; Zhang, X.; Luo, Z.; Huang, L.; Chen, X.; Li, Y. De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of cSSR markers in sweet potato (Ipomoea batatas). BMC Genom. 2010, 11, 726. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, J.A.; Yang, S.S.; Miller, S.S.; Bucciarelli, B.; Liu, J.; Rydeen, A.; Bozsoki, Z.; Uhde-Stone, C.; Tu, Z.J.; Allan, D.; et al. An RNA-Seq transcriptome analysis of orthophosphate-deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol. 2013, 161, 705–724. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raktima, R.; Qiandong, Z.; Zehua, C.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Byeong-ha, L.; David, A.H.; Zhu, J.K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 2005, 17, 3155–3175. [Google Scholar]
- Kyonoshin, M.; Sakuma, Y.; Kasuga, M.; Ito, Y.; Seki, M.; Goda, H.; Shimada, Y.; Yoshida, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 2004, 38, 982–993. [Google Scholar]
- Knight, M.R.; Knight, H. Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol. 2012, 195, 737–751. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Zarka, D.G.; Stockinger, E.J.; Salazar, M.P.; Houghton, J.M.; Thomashow, M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998, 16, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Fowler, S.G.; Cheng, H.; Lou, Y.; Rhee, S.Y.; Stockinger, E.J.; Thomashow, M.F. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J. 2004, 39, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.T.; Zarka, D.G.; Van-Buskirk, H.A.; Fowler, S.G.; Thomashow, M.F. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005, 41, 195–211. [Google Scholar] [CrossRef]
- Novillo, F.; Medina, J.; Salinas, J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA 2007, 104, 21002–21007. [Google Scholar] [CrossRef] [PubMed]
- Fowler, S.; Thomashow, M.F. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 2002, 14, 1675–1690. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, K.; Bucher, P. The FHA domain: A putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem. Sci. 1995, 20, 347–349. [Google Scholar] [CrossRef]
- Durocher, D.; Jackson, S.P. The FHA domain. FEBS Lett. 2002, 513, 58–66. [Google Scholar] [CrossRef]
- Liscum, E.; Reed, J.W. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 2002, 49, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 2004, 16, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Wang, X.J.; Hagen, G.; Guilfoyle, T.J. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 2001, 13, 2809–2822. [Google Scholar] [CrossRef] [PubMed]
- Allison, L.A. The role of sigma factors in plastid transcription. Biochimie 2000, 82, 537–548. [Google Scholar] [CrossRef]
- Paget, M.S.; Helmann, J.D. The sigma70 family of sigma factors. Genome Biol. 2003, 4, 203. [Google Scholar] [CrossRef] [PubMed]
- Sriraman, P.; Silhavy, D.; Maliga, P. Transcription from heterologous rRNA operon promoters in chloroplasts reveals requirement for specific activating factors. Plant Physiol. 1998, 117, 1495–1499. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wei, W.; Pang, X.; Wang, X.; Zhang, H.; Dong, B.; Xing, Y.; Li, X.; Wang, M. Comparative transcriptome profiling of a desert evergreen shrub, Ammopiptanthus mongolicus, in response to drought and cold stresses. BMC Genom. 2014, 15, 671. [Google Scholar] [CrossRef] [PubMed]
- Doerks, T.; Copley, R.; Bork, P. DDT—a novel domain in different transcription and chromosome remodeling factors. Trends Biochem. Sci. 2001, 26, 145–146. [Google Scholar] [CrossRef]
- Guo, X.; Xu, S.; Chong, K. Cold signal shuttles from membrane to nucleus. Mol. Cell 2017, 66, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, F.; Yan, P.; Jing, W.; Zhang, C.; Kudla, J.; Zhang, W. A phosphoinositide-specific phospholipase C pathway elicits stress-induced Ca2+ signals and confers salt tolerance to rice. New Phytol. 2017, 214, 1172–1187. [Google Scholar] [CrossRef] [PubMed]
- Tarutani, Y.; Morimoto, T.; Sasaki, A.; Yasuda, M.; Nakashita, H.; Yoshida, S.; Yamaguchi, I.; Suzuki, Y. Molecular characterization of two highly homologous receptor-like kinase genes, RLK902 and RKL1, in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2004, 68, 1935–1941. [Google Scholar] [CrossRef] [PubMed]
- Tarutani, Y.; Sasaki, A.; Yasuda, M.; Nakashita, H.; Yoshida, S.; Yamaguchi, I.; Suzuki, Y. Identification of three clones which commonly interact with the kinase domains of highly homologous two receptor-like kinases, RLK902 and RKL1. Biosci. Biotechnol. Biochem. 2004, 68, 2581–2587. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.W.; Jon, J.H.; Kwak, J.M.; Nam, H.G. Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopsis thaliana. Plant Physiol. 1997, 113, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Maruyama, K.; Seki, M.; Satou, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell. 2005, 17, 1105–1119. [Google Scholar] [CrossRef] [PubMed]
- Van-Helvoort, A.; Smith, A.J.; Sprong, H.; Fritzsche, I.; Schinkel, A.H.; Borst, P.; Van-Meer, G. MDR1 P-Glycoprotein is a lipid translocase of broad specificity, while MDR3 P-glycoprotein specifically translocates phosphatidylcholine. Cell 1996, 87, 507–517. [Google Scholar] [CrossRef]
- Higgins, C.F. ABC TRANSPORTERS: From microorganisms to man. Annu. Rev. Cell Biol. 1992, 8, 67–113. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, Y.; Liu, X.; Huang, J.; Wang, Q.; Gu, J.; Lu, Y. Transcriptome profiling of the cold response and signaling pathways in Lilium lancifolium. BMC Genom. 2014, 15, 203. [Google Scholar] [CrossRef] [PubMed]





| Sample | Condition | Clean Reads | Sequence Length (bp) | Total Bases (G) | %GC |
|---|---|---|---|---|---|
| ×3 | 20 °C → 4 °C (7 days) | 35,597,819 | 101 | 3.5 | 45.56 |
| 35,597,819 | 101 | 3.5 | 45.56 | ||
| ×6 | 20 °C → −2 °C (12 h) | 42,896,402 | 101 | 4.3 | 45.28 |
| 42,896,402 | 101 | 4.3 | 45.28 | ||
| ×5 | 20 °C → 4 °C (7 days) → 0 °C (24 h) → −2 °C (24 h) | 33,720,829 | 101 | 3.4 | 45.45 |
| 33,720,829 | 101 | 3.4 | 45.45 | ||
| ×4 | 20 °C → 4 °C (7 days) → 0 °C (24 h) | 40,779,352 | 101 | 4.1 | 44.73 |
| 40,779,352 | 101 | 4.1 | 44.73 | ||
| ×1 | 20 °C | 38,880,446 | 101 | 3.9 | 47.78 |
| 38,880,446 | 101 | 3.9 | 47.78 | ||
| ×2 | 20 °C → 4 °C (24 h) | 41,863,445 | 101 | 4.2 | 46.32 |
| 41,863,445 | 101 | 4.2 | 46.32 | ||
| Assembly Statistic (Transcripts) | |||||
| Total Sequences | Min Length | Average Length | Max Length | N50 | %GC |
| 199,758 | 201 | 711.37 | 15,818 | 1099 | 39.05 |
| Category | 4 °C (24 h) ×2 | 4 °C (7 days) ×3 | 0 °C (24 h) ×4 | −2 °C (24 h) ×5 | −2 °C (12 h) ×6 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | |
| Protein kinase | 11 | 15 | 17 | 7 | 11 | 12 | 27 | 23 | 27 | 33 |
| Protein phosphatase | 9 | 2 | 14 | 1 | 13 | 3 | 22 | 8 | 15 | 5 |
| Receptor-like kinase | 12 | 24 | 8 | 41 | 21 | 23 | 32 | 86 | 16 | 42 |
| Histidine kinase | 2 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| Rhythm signal | 2 | 3 | 1 | 1 | 9 | 1 | 14 | 4 | 2 | 2 |
| GTP-related | 0 | 1 | 0 | 3 | 0 | 0 | 1 | 7 | 0 | 8 |
| Lipid-signal | 3 | 3 | 3 | 1 | 3 | 4 | 9 | 7 | 0 | 11 |
| Calcium-related | 1 | 4 | 0 | 1 | 3 | 6 | 14 | 12 | 0 | 5 |
| Category | 4 °C (24 h) ×2 | 4 °C (7 days) ×3 | 0 °C (24 h) ×4 | −2 °C (24 h) ×5 | −2 °C (12 h) ×6 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | |
| Ion-related | 5 | 7 | 5 | 9 | 12 | 5 | 14 | 19 | 12 | 15 |
| Carbon-related | 1 | 1 | 5 | 3 | 3 | 2 | 1 | 8 | ||
| Organ-related | 3 | 1 | 2 | 3 | 3 | 5 | 8 | 4 | 3 | |
| ABC family | 3 | 5 | 5 | 2 | 9 | 8 | 4 | 9 | ||
| Nucleotide | 2 | 1 | 3 | 3 | 1 | 3 | 4 | 4 | 4 | |
| Auxin-related | 2 | 1 | 3 | 3 | 1 | 3 | 4 | |||
| Amino acid | 5 | 3 | 5 | 3 | 5 | 3 | 11 | 7 | 6 | 9 |
| Aquaporin | 1 | 1 | 1 | 2 | 2 | 3 | 1 | |||
| MATE efflux family | 1 | 1 | 3 | 2 | 1 | 5 | 6 | 3 | ||
| Proton pump | 1 | 1 | 4 | 3 | 2 | 4 | 7 | 3 | 3 | |
| NRT1/PTR family | 2 | 1 | 3 | 2 | 4 | 3 | 6 | 8 | 4 | 6 |
| Inorganic salt | 2 | 5 | 2 | 7 | 3 | 1 | ||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Liu, H.; Xia, W.; Mu, J.; Feng, Y.; Liu, R.; Yan, P.; Wang, A.; Lin, Z.; Guo, Y.; et al. De Novo Transcriptome Sequencing and the Hypothetical Cold Response Mode of Saussurea involucrata in Extreme Cold Environments. Int. J. Mol. Sci. 2017, 18, 1155. https://doi.org/10.3390/ijms18061155
Li J, Liu H, Xia W, Mu J, Feng Y, Liu R, Yan P, Wang A, Lin Z, Guo Y, et al. De Novo Transcriptome Sequencing and the Hypothetical Cold Response Mode of Saussurea involucrata in Extreme Cold Environments. International Journal of Molecular Sciences. 2017; 18(6):1155. https://doi.org/10.3390/ijms18061155
Chicago/Turabian StyleLi, Jin, Hailiang Liu, Wenwen Xia, Jianqiang Mu, Yujie Feng, Ruina Liu, Panyao Yan, Aiying Wang, Zhongping Lin, Yong Guo, and et al. 2017. "De Novo Transcriptome Sequencing and the Hypothetical Cold Response Mode of Saussurea involucrata in Extreme Cold Environments" International Journal of Molecular Sciences 18, no. 6: 1155. https://doi.org/10.3390/ijms18061155
APA StyleLi, J., Liu, H., Xia, W., Mu, J., Feng, Y., Liu, R., Yan, P., Wang, A., Lin, Z., Guo, Y., Zhu, J., & Chen, X. (2017). De Novo Transcriptome Sequencing and the Hypothetical Cold Response Mode of Saussurea involucrata in Extreme Cold Environments. International Journal of Molecular Sciences, 18(6), 1155. https://doi.org/10.3390/ijms18061155






