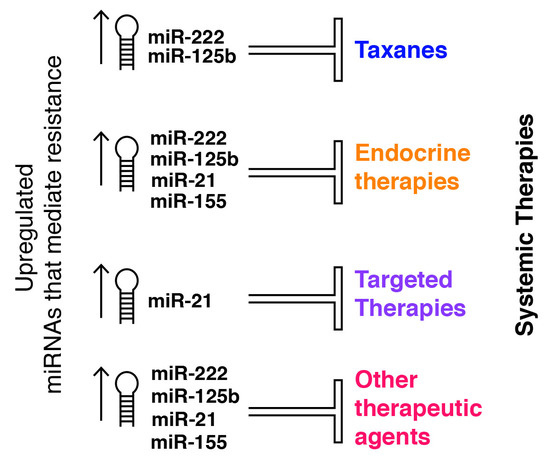
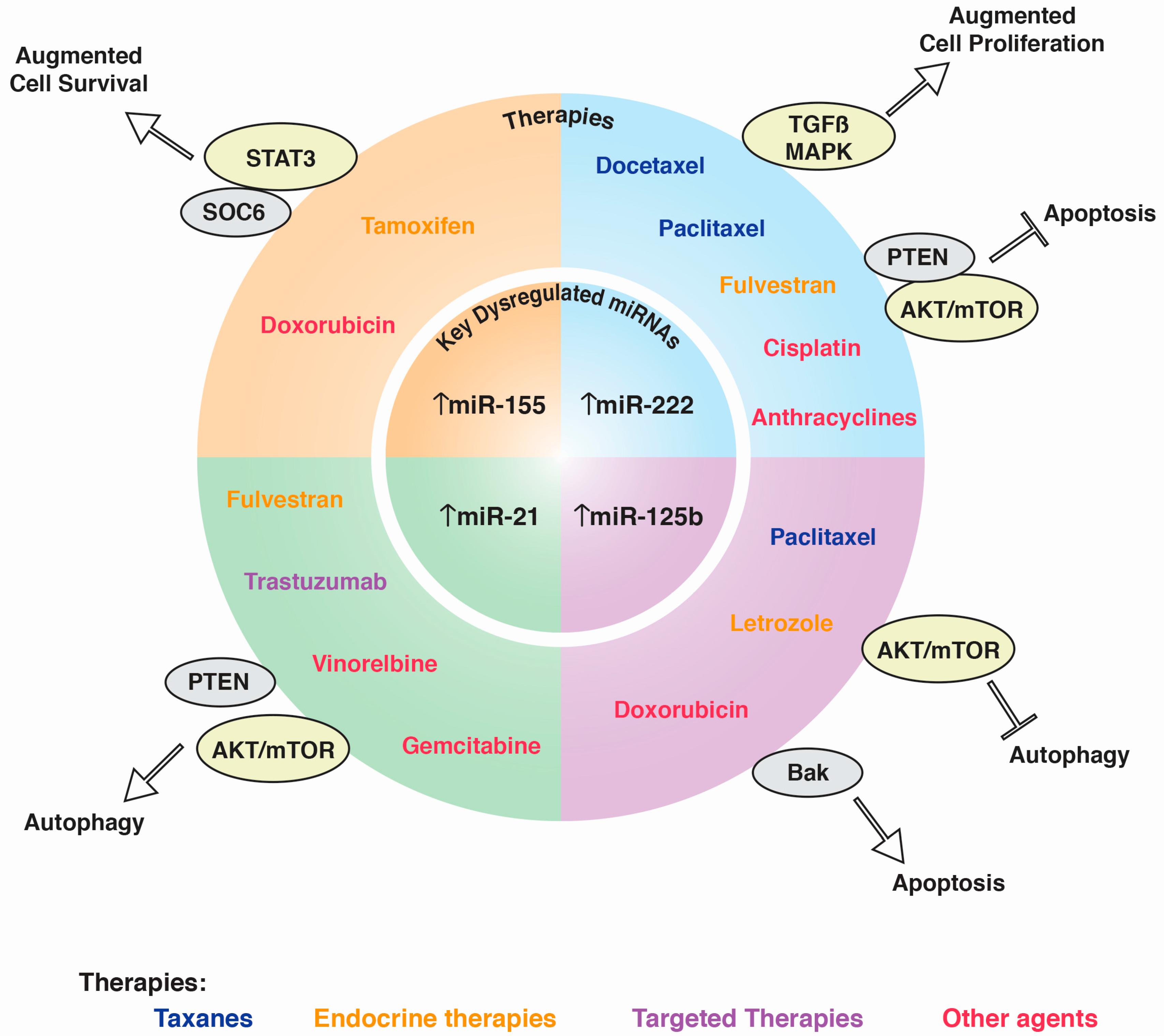
Micro-RNAs as Potential Predictors of Response to Breast Cancer Systemic Therapy: Future Clinical Implications
Abstract
:1. Background
2. Main Text
2.1. miRNAs in Taxane Resistance (Paclitaxel, Docetaxel)
2.2. Paclitaxel
2.3. Docetaxel
2.4. miRNAs in Endocrine Therapy
2.5. Aromatase Inhibitors
2.6. miRNAs in SERM Therapy
2.7. miRNAs in Targeted Therapies
2.7.1. Trastuzumab
2.7.2. Lapatinib
2.8. Other Agents
2.8.1. Anthracyclines
2.8.2. Cisplatin
2.8.3. Capecitabine, Gemcitabine, and Vinorelbine
3. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BC | Breast Cancer |
| miRNA | MicroRNA |
| Bak1 | Pro-apoptotic Bcl2 antagonist killer 1 |
| DAPK2 | Death-Associated kinase 2 |
| TSP-1 | Thrombospondin-1 |
| MCL-1 | Myeloid cell leukemia |
| EIF4E | Eukaryotic translation initiation factor 4E |
| CP110 | Centriolar coiled-coil protein 110 |
| APC4 | Anaphase promoting complex subunit 4 |
| HSPG2 | Heparin sulfate proteoglycan 2 |
| CCND1 | Cyclin D1 |
| PTEN | Phosphatase and tensin homolog |
| ERα | Estrogen receptor α |
| SERDs | Selective estrogen receptor downregulators |
| SERMs | Selective ER modulators |
| TCGA | The cancer genome atlas |
| ASCO | American society of clinical oncology |
| GEMIN4 | Gem nuclear organelle associated protein 4 |
| BMP7 | Bone morphogenetic protein 7 |
| UCP2 | Uncoupling protein 2 |
| HDAC4 | Histone deacetylase |
| SOC6 | Suppressor of cytokine signaling 6 |
| HER2 | Human epidermal growth factor receptor-2 |
| EMT | Epithelial to mesenchymal transition |
| IGF1R | Insulin-like growth factor-1 receptor |
| TKI | Tyrosine kinase inhibitor |
| FDA | Food and drug administration |
| CCNJ | Cyclin J |
| FUBP1 | Far upstream element binding protein 1 |
| TNBC | Triple negative breast cancer |
| IL-6 | Interleukine-6 |
References
- Jemal, A.; Siegel, R.; Xu, J.; Ward, E. Cancer statistics, 2010. Cancer J. Clin. 2010, 60, 277–300. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.; Harbeck, N.; Barrios, C.H.; Bergh, J.; Cortés, J.; El Saghir, N.; Francis, P.A.; Hudis, C.A.; Ohno, S.; Partridge, A.H.; et al. Research needs in breast cancer. Ann. Oncol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2016. [Google Scholar] [CrossRef]
- Marquette, C.; Nabell, L. Chemotherapy-resistant metastatic breast cancer. Curr. Treat. Opt. Oncol. 2012, 13, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jeon, K.; Lee, J.-T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Cullen, B.R. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004, 32, 4776–4785. [Google Scholar] [CrossRef] [PubMed]
- Lingel, A.; Simon, B.; Izaurralde, E.; Sattler, M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat. Struct. Mol. Biol. 2004, 11, 576–577. [Google Scholar] [CrossRef] [PubMed]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Corsini, L.R.; Bronte, G.; Terrasi, M.; Amodeo, V.; Fanale, D.; Fiorentino, E.; Cicero, G.; Bazan, V.; Russo, A. The role of microRNAs in cancer: Diagnostic and prognostic biomarkers and targets of therapies. Expert Opin. Ther. Targets 2012, 16, S103–S109. [Google Scholar] [CrossRef] [PubMed]
- O’Day, E.; Lal, A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 2010, 12, 201. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.T.; Nagai, S.; Chen, B.K.; Qureshi, Z.P.; Lebby, A.A.; Kessler, S.; Georgantopoulos, P.; Raisch, D.W.; Sartor, O.; Hermanson, T.; et al. Generic oncology drugs: Are they all safe? Lancet Oncol. 2016, 17, e493–e501. [Google Scholar] [CrossRef]
- Sparano, J.A. Taxanes for breast cancer: An evidence-based review of randomized phase II and phase III trials. Clin. Breast Cancer 2000. [Google Scholar] [CrossRef] [PubMed]
- Crown, J.; O’Leary, M.; Ooi, W.-S. Docetaxel and paclitaxel in the treatment of breast cancer: A review of clinical experience. Oncologist 2004, 9, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.E.; Erban, J.; Overmoyer, B.; Budd, G.T.; Hutchins, L.; Lower, E.; Laufman, L.; Sundaram, S.; Urba, W.J.; Pritchard, K.I.; et al. Randomized phase III study of docetaxel compared with paclitaxel in metastatic breast cancer. J. Clin. Oncol. 2005, 23, 5542–5551. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.-X.; Shen, Z.; Lin, F.; Sun, Y.; Min, D.; Tang, L.-N.; He, A.-N.; Yao, Y. Paclitaxel-based versus docetaxel-based regimens in metastatic breast cancer: A systematic review and meta-analysis of randomized controlled trials. Curr. Med. Res. Opin. 2013, 29, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Del Mastro, L.; Fabi, A.; Mansutti, M.; de Laurentiis, M.; Durando, A.; Merlo, D.F.; Bruzzi, P.; La Torre, I.; Ceccarelli, M.; Kazeem, G.; et al. Randomised phase III open-label trial of first-line treatment with gemcitabine in association with docetaxel or paclitaxel in women with metastatic breast cancer: A comparison of different schedules and treatments. BMC Cancer 2013, 13, 164. [Google Scholar] [CrossRef] [PubMed]
- Belfiglio, M.; Fanizza, C.; Tinari, N.; Ficorella, C.; Iacobelli, S.; Natoli, C. Consorzio Interuniversitario Nazionale per la Bio-Oncologia (CINBO) Meta-analysis of phase III trials of docetaxel alone or in combination with chemotherapy in metastatic breast cancer. J. Cancer Res. Clin. Oncol. 2012, 138, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Lück, H.-J.; Lübbe, K.; Reinisch, M.; Maass, N.; Feisel-Schwickardi, G.; Tomé, O.; Janni, W.; Aydogdu, M.; Neunhöffer, T.; Ober, A.; et al. Phase III study on efficacy of taxanes plus bevacizumab with or without capecitabine as first-line chemotherapy in metastatic breast cancer. Breast Cancer Res. Treat. 2015, 149, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.M.; Opdenaker, L.M.; Flynn, D.; Sims-Mourtada, J. Wound healing and cancer stem cells: Inflammation as a driver of treatment resistance in breast cancer. Cancer Growth Metastasis 2015, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lv, K.; Liu, L.; Wang, L.; Yu, J.; Liu, X.; Cheng, Y.; Dong, M.; Teng, R.; Wu, L.; Fu, P.; et al. Lin28 mediates paclitaxel resistance by modulating p21, Rb and Let-7a miRNA in breast cancer cells. PLoS ONE 2012, 7, e40008. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Liu, Z.; Zhao, Y.; Ding, Y.; Liu, H.; Xi, Y.; Xiong, W.; Li, G.; Lu, J.; Fodstad, O.; et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J. Biol. Chem. 2010, 285, 21496–21507. [Google Scholar] [CrossRef] [PubMed]
- Su, C.-M.; Wang, M.-Y.; Hong, C.-C.; Chen, H.-A.; Su, Y.-H.; Wu, C.-H.; Huang, M.-T.; Chang, Y.-W.; Jiang, S.-S.; Sung, S.-Y.; et al. miR-520h is crucial for DAPK2 regulation and breast cancer progression. Oncogene 2016, 35, 1134–1142. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Li, J.-Y.; Guo, J.; Li, P.-S.; Zhang, W.-H. Influence of miR-451 on Drug Resistances of Paclitaxel-Resistant Breast Cancer Cell Line. Med. Sci. Monit. 2015, 21, 3291–3297. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhao, R.; He, Y.; Fu, X.; Fu, L.; Zhu, Z.; Fu, L.; Dong, J.-T. MicroRNA 100 sensitizes luminal A breast cancer cells to paclitaxel treatment in part by targeting mTOR. Oncotarget 2016, 7, 5702–5714. [Google Scholar] [CrossRef] [PubMed]
- Sha, L.-Y.; Zhang, Y.; Wang, W.; Sui, X.; Liu, S.-K.; Wang, T.; Zhang, H. miR-18a upregulation decreases Dicer expression and confers paclitaxel resistance in triple negative breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2201–2208. [Google Scholar] [PubMed]
- Tsang, W.P.; Kwok, T.T. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis 2008, 13, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.-Y.; Liang, X.-S.; Liu, Y.; Wang, C.-Y.; Pang, D. Decrease of let-7f in low-dose metronomic Paclitaxel chemotherapy contributed to upregulation of thrombospondin-1 in breast cancer. Int. J. Biol. Sci. 2015, 11, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tang, H.; Chen, J.; Song, C.; Yang, L.; Liu, P.; Wang, N.; Xie, X.; Lin, X.; Xie, X. MicroRNA-101 inhibits cell progression and increases paclitaxel sensitivity by suppressing MCL-1 expression in human triple-negative breast cancer. Oncotarget 2015, 6, 20070–20083. [Google Scholar] [CrossRef] [PubMed]
- Nabholtz, J.-M.; Gligorov, J. Docetaxel in the treatment of breast cancer: Current experience and future prospects. Expert Rev. Anticancer Ther. 2005, 5, 613–633. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.-S.; Qiu, W.-S.; Yao, R.-Y.; Zhang, Q.; Zhuang, L.-K.; Zhou, F.; Sun, L.-B.; Yue, L. miR-141 confers docetaxel chemoresistance of breast cancer cells via regulation of EIF4E expression. Oncol. Rep. 2015, 33, 2504–2512. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Wei, Y.; Li, M.; Yu, S.; Ye, M.; Zhang, H.; Chen, S.; Liu, W.; Zhang, J. miR-129-3p promotes docetaxel resistance of breast cancer cells via CP110 inhibition. Sci. Rep. 2015, 5, 15424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhong, S.; Xu, Y.; Yu, D.; Ma, T.; Chen, L.; Zhao, Y.; Chen, X.; Yang, S.; Wu, Y.; et al. MicroRNA-3646 Contributes to Docetaxel Resistance in Human Breast Cancer Cells by GSK-3β/β-Catenin Signaling Pathway. PLoS ONE 2016, 11, e0153194. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Chen, W.; Zhong, S.; Zhang, J.; Ma, T.; Ji, H.; Lv, M.; Tang, J.; Zhao, J. MicroRNA-452 contributes to the docetaxel resistance of breast cancer cells. Tumour Biol. 2014, 35, 6327–6334. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Li, S.; Cui, X.; Lv, X.; Jiao, Y.; Yu, F.; Yao, H.; Song, E.; Chen, Y.; Wang, M.; et al. The overexpression of hypomethylated miR-663 induces chemotherapy resistance in human breast cancer cells by targeting heparin sulfate proteoglycan 2 (HSPG2). J. Biol. Chem. 2013, 288, 10973–10985. [Google Scholar] [CrossRef] [PubMed]
- Kastl, L.; Brown, I.; Schofield, A.C. miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res. Treat. 2012, 131, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Sun, D.-W.; Mao, L.; Zhang, J.; Jiang, L.-H.; Li, J.; Wu, Y.; Ji, H.; Chen, W.; Wang, J.; et al. miR-139-5p inhibits the biological function of breast cancer cells by targeting Notch1 and mediates chemosensitivity to docetaxel. Biochem. Biophys. Res. Commun. 2015, 465, 702–713. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Yan, X.; Zhang, G.; Zhao, W.; Jiao, S. MicroRNA-205 increases the sensitivity of docetaxel in breast cancer. Oncol. Lett. 2016, 11, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lv, Y.-G.; Yan, C.-Y.; Yi, J.; Ling, R. Enforced expression of hsa-miR-125a-3p in breast cancer cells potentiates docetaxel sensitivity via modulation of BRCA1 signaling. Biochem. Biophys. Res. Commun. 2016, 479, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Li, W.; Chen, Z.; Xu, J.; Zhao, J. miR-222 and miR-29a contribute to the drug-resistance of breast cancer cells. Gene 2013, 531, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-X.; Cai, Y.-Q.; Lv, M.-M.; Chen, L.; Zhong, S.-L.; Ma, T.-F.; Zhao, J.-H.; Tang, J.-H. Exosomes from docetaxel-resistant breast cancer cells alter chemosensitivity by delivering microRNAs. Tumour Biol. 2014, 35, 9649–9659. [Google Scholar] [CrossRef] [PubMed]
- Stanford, J.L.; Szklo, M.; Brinton, L.A. Estrogen receptors and breast cancer. Epidemiol. Rev. 1986, 8, 42–59. [Google Scholar] [CrossRef] [PubMed]
- Beato, M.; Herrlich, P.; Schütz, G. Steroid hormone receptors: Many actors in search of a plot. Cell 1995, 83, 851–857. [Google Scholar] [CrossRef]
- Tsai, M.J.; O’Malley, B.W. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 1994, 63, 451–486. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Green, S.; Stack, G.; Berry, M.; Jin, J.R.; Chambon, P. Functional domains of the human estrogen receptor. Cell 1987, 51, 941–951. [Google Scholar] [CrossRef]
- Regan, M.M.; Neven, P.; Giobbie-Hurder, A.; Goldhirsch, A.; Ejlertsen, B.; Mauriac, L.; Forbes, J.F.; Smith, I.; Láng, I.; Wardley, A.; et al. Assessment of letrozole and tamoxifen alone and in sequence for postmenopausal women with steroid hormone receptor-positive breast cancer: The BIG 1–98 randomised clinical trial at 81 years median follow-up. Lancet Oncol. 2011, 12, 1101–1108. [Google Scholar] [CrossRef]
- Johnston, S.R.D.; Martin, L.-A.; Head, J.; Smith, I.; Dowsett, M. Aromatase inhibitors: Combinations with fulvestrant or signal transduction inhibitors as a strategy to overcome endocrine resistance. J. Steroid Biochem. Mol. Biol. 2005, 95, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-Z.; Xu, Q.-N.; Wang, H.-B.; Li, X.-Y. Fulvestrant plus targeted agents versus fulvestrant alone for treatment of hormone-receptor positive advanced breast cancer progressed on previous endocrine therapy: A meta-analysis of randomized controlled trials. Breast Cancer 2017, 24, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.L. The Long and Short of Tamoxifen Therapy: A Review of the ATLAS Trial. J. Adv. Pract. Oncol. 2014, 5, 57–60. [Google Scholar] [PubMed]
- Gilani, R.A.; Kazi, A.A.; Shah, P.; Schech, A.J.; Chumsri, S.; Sabnis, G.; Jaiswal, A.K.; Brodie, A.H. The importance of HER2 signaling in the tumor-initiating cell population in aromatase inhibitor-resistant breast cancer. Breast Cancer Res. Treat. 2012, 135, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Burris, H.A. Overcoming acquired resistance to anticancer therapy: Focus on the PI3K/AKT/mTOR pathway. Cancer Chemother. Pharmacol. 2013, 71, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zou, Z.; Nie, P.; Kou, X.; Wu, B.; Wang, S.; Song, Z.; He, J. Downregulation of microRNA-27b-3p enhances tamoxifen resistance in breast cancer by increasing NR5A2 and CREB1 expression. Cell Death Dis. 2016, 7, e2454. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-R.; Song, Y.; Wan, L.-H.; Zhang, Y.-Y.; Zhou, L.-M. Over-expression of miR-451a can enhance the sensitivity of breast cancer cells to tamoxifen by regulating 14-3-3ζ, estrogen receptor α, and autophagy. Life Sci. 2016, 149, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Vilquin, P.; Donini, C.F.; Villedieu, M.; Grisard, E.; Corbo, L.; Bachelot, T.; Vendrell, J.A.; Cohen, P.A. MicroRNA-125b upregulation confers aromatase inhibitor resistance and is a novel marker of poor prognosis in breast cancer. Breast Cancer Res. 2015, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Bailey, S.T.; Westerling, T.; Brown, M. Loss of estrogen-regulated microRNA expression increases HER2 signaling and is prognostic of poor outcome in luminal breast cancer. Cancer Res. 2015, 75, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Luo, A.; Liu, Y.; Wang, S.; Li, Y.; Shi, W.; Liu, Z.; Qu, X. MiR-214 increases the sensitivity of breast cancer cells to tamoxifen and fulvestrant through inhibition of autophagy. Mol. Cancer 2015, 14, 208. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Di Leva, G.; Li, M.; Fang, F.; Devlin, C.; Hartman-Frey, C.; Burow, M.E.; Ivan, M.; Croce, C.M.; Nephew, K.P. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene 2011, 30, 1082–1097. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, R.; Shi, W.; Jiang, T.; Wang, Y.; Li, C.; Qu, X. Silencing of microRNA-21 confers the sensitivity to tamoxifen and fulvestrant by enhancing autophagic cell death through inhibition of the PI3K-AKT-mTOR pathway in breast cancer cells. Biomed. Pharmacother. 2016, 77, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Maximov, P.Y.; Lee, T.M.; Jordan, V.C. The discovery and development of selective estrogen receptor modulators (SERMs) for clinical practice. Curr. Clin. Pharmacol. 2013, 8, 135–155. [Google Scholar] [CrossRef] [PubMed]
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG); Davies, C.; Godwin, J.; Gray, R.; Clarke, M.; Cutter, D.; Darby, S.; McGale, P.; Pan, H.C.; Taylor, C.; et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: Patient-level meta-analysis of randomised trials. Lancet 2011, 378, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-González, F.G.; Sieuwerts, A.M.; Smid, M.; Look, M.P.; Meijer-van Gelder, M.E.; de Weerd, V.; Sleijfer, S.; Martens, J.W.M.; Foekens, J.A. MicroRNA-30c expression level is an independent predictor of clinical benefit of endocrine therapy in advanced estrogen receptor positive breast cancer. Breast Cancer Res. Treat. 2011, 127, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, R.; Achinger-Kawecka, J.; Winter, S.; Fritz, P.; Lo, W.-Y.; Schroth, W.; Brauch, H. Increased expression of miR-126 and miR-10a predict prolonged relapse-free time of primary oestrogen receptor-positive breast cancer following tamoxifen treatment. Eur. J. Cancer 2013, 49, 3598–3608. [Google Scholar] [CrossRef] [PubMed]
- Rothé, F.; Ignatiadis, M.; Chaboteaux, C.; Haibe-Kains, B.; Kheddoumi, N.; Majjaj, S.; Badran, B.; Fayyad-Kazan, H.; Desmedt, C.; Harris, A.L.; et al. Global microRNA expression profiling identifies miR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS ONE 2011, 6, e20980. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Ginnebaugh, K.R.; Yin, S.; Bollig-Fischer, A.; Reddy, K.B.; Sarkar, F.H. Functional role of miR-10b in tamoxifen resistance of ER-positive breast cancer cells through down-regulation of HDAC4. BMC Cancer 2015, 15, 540. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Wang, Y.; Wang, C.-X.; Yin, M.; Liu, H.-L.; Chen, J.-P.; Han, J.-Q.; Wang, W.-B. miRNA-155 mediates TAM resistance by modulating SOCS6-STAT3 signalling pathway in breast cancer. Am. J. Transl. Res. 2015, 7, 2115–2126. [Google Scholar] [PubMed]
- Vu, T.; Claret, F.X. Trastuzumab: Updated mechanisms of action and resistance in breast cancer. Front. Oncol. 2012, 2, 62. [Google Scholar] [CrossRef] [PubMed]
- Slamon, D.J.; Leyland-Jones, B.; Shak, S.; Fuchs, H.; Paton, V.; Bajamonde, A.; Fleming, T.; Eiermann, W.; Wolter, J.; Pegram, M.; et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 2001, 344, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, R.L.; Paw, I.; Dewhirst, M.W.; Lo, H.-W. Akt phosphorylates and activates HSF-1 independent of heat shock, leading to Slug overexpression and epithelial-mesenchymal transition (EMT) of HER2-overexpressing breast cancer cells. Oncogene 2015, 34, 546–557. [Google Scholar] [CrossRef] [PubMed]
- De Mattos-Arruda, L.; Bottai, G.; Nuciforo, P.G.; Di Tommaso, L.; Giovannetti, E.; Peg, V.; Losurdo, A.; Pérez-Garcia, J.; Masci, G.; Corsi, F.; et al. MicroRNA-21 links epithelial-to-mesenchymal transition and inflammatory signals to confer resistance to neoadjuvant trastuzumab and chemotherapy in HER2-positive breast cancer patients. Oncotarget 2015, 6, 37269–37280. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Yuan, P.; Zhao, Z.T.; Yang, Z.; Wang, T.; Zhao, J.D.; Luo, Y.; Ma, F.; Wang, J.Y.; Fan, Y.; et al. A miRNA-based signature predicts development of disease recurrence in HER2 positive breast cancer after adjuvant trastuzumab-based treatment. Sci. Rep. 2016, 6, 33825. [Google Scholar] [CrossRef] [PubMed]
- Jung, E.-J.; Santarpia, L.; Kim, J.; Esteva, F.J.; Moretti, E.; Buzdar, A.U.; di Leo, A.; Le, X.-F.; Bast, R.C.; Park, S.-T.; et al. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer 2012, 118, 2603–2614. [Google Scholar] [CrossRef] [PubMed]
- Crosby, M.E.; Kulshreshtha, R.; Ivan, M.; Glazer, P.M. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009, 69, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zi, X.; Zhao, Y.; Mascarenhas, D.; Pollak, M. Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (Herceptin). J. Natl. Cancer Inst. 2001, 93, 1852–1857. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.-M.; Zhu, H.-Y.; Bai, W.-D.; Wang, T.; Wang, L.; Chen, Y.; Yang, A.-G.; Jia, L.-T. Epigenetic silencing of miR-375 induces trastuzumab resistance in HER2-positive breast cancer by targeting IGF1R. BMC Cancer 2014, 14, 134. [Google Scholar] [CrossRef] [PubMed]
- Geyer, C.E.; Forster, J.; Lindquist, D.; Chan, S.; Romieu, C.G.; Pienkowski, T.; Jagiello-Gruszfeld, A.; Crown, J.; Chan, A.; Kaufman, B.; et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N. Engl. J. Med. 2006, 355, 2733–2743. [Google Scholar] [CrossRef] [PubMed]
- Johnston, S.; Pippen, J.; Pivot, X.; Lichinitser, M.; Sadeghi, S.; Dieras, V.; Gomez, H.L.; Romieu, G.; Manikhas, A.; Kennedy, M.J.; et al. Lapatinib combined with letrozole versus letrozole and placebo as first-line therapy for postmenopausal hormone receptor-positive metastatic breast cancer. J. Clin. Oncol. 2009, 27, 5538–5546. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Mullin, R.J.; Keith, B.R.; Liu, L.-H.; Ma, H.; Rusnak, D.W.; Owens, G.; Alligood, K.J.; Spector, N.L. Anti-tumor activity of GW572016: A dual tyrosine kinase inhibitor blocks EGF activation of EGFR/erbB2 and downstream Erk1/2 and AKT pathways. Oncogene 2002, 21, 6255–6263. [Google Scholar] [CrossRef] [PubMed]
- Scaltriti, M.; Rojo, F.; Ocaña, A.; Anido, J.; Guzman, M.; Cortes, J.; di Cosimo, S.; Matias-Guiu, X.; Ramon y Cajal, S.; Arribas, J.; et al. Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. J. Natl. Cancer Inst. 2007, 99, 628–638. [Google Scholar] [CrossRef] [PubMed]
- de Azambuja, E.; Holmes, A.P.; Piccart-Gebhart, M.; Holmes, E.; di Cosimo, S.; Swaby, R.F.; Untch, M.; Jackisch, C.; Lang, I.; Smith, I.; et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): Survival outcomes of a randomised, open-label, multicentre, phase III trial and their association with pathological complete response. Lancet Oncol. 2014, 15, 1137–1146. [Google Scholar] [CrossRef]
- Guan, Z.; Xu, B.; DeSilvio, M.L.; Shen, Z.; Arpornwirat, W.; Tong, Z.; Lorvidhaya, V.; Jiang, Z.; Yang, J.; Makhson, A.; et al. Randomized trial of lapatinib versus placebo added to paclitaxel in the treatment of human epidermal growth factor receptor 2-overexpressing metastatic breast cancer. J. Clin. Oncol. 2013, 31, 1947–1953. [Google Scholar] [CrossRef] [PubMed]
- Esteva, F.J.; Yu, D.; Hung, M.-C.; Hortobagyi, G.N. Molecular predictors of response to trastuzumab and lapatinib in breast cancer. Nat. Rev. Clin. Oncol. 2010, 7, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Venturutti, L.; Cordo Russo, R.I.; Rivas, M.A.; Mercogliano, M.F.; Izzo, F.; Oakley, R.H.; Pereyra, M.G.; de Martino, M.; Proietti, C.J.; Yankilevich, P.; et al. miR-16 mediates trastuzumab and lapatinib response in ErbB-2-positive breast and gastric cancer via its novel targets CCNJ and FUBP1. Oncogene 2016, 35, 6189–6202. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, C.; Rani, S.; Breslin, S.; Gogarty, M.; Ghobrial, I.M.; Crown, J.; O’Driscoll, L. miR-630 targets IGF1R to regulate response to HER-targeting drugs and overall cancer cell progression in HER2 over-expressing breast cancer. Mol. Cancer 2014, 13, 71. [Google Scholar] [CrossRef] [PubMed]
- Siziopikou, K.P.; Cobleigh, M. The basal subtype of breast carcinomas may represent the group of breast tumors that could benefit from EGFR-targeted therapies. Breast Edinb. Scotl. 2007, 16, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Press, M.F.; Dering, J.; Arbushites, M.; Koehler, M.; Oliva, C.; Williams, L.S.; di Leo, A. Estrogen receptor, progesterone receptor, human epidermal growth factor receptor 2 (HER2), and epidermal growth factor receptor expression and benefit from lapatinib in a randomized trial of paclitaxel with lapatinib or placebo as first-line treatment in HER2-negative or unknown metastatic breast cancer. J. Clin. Oncol. 2009, 27, 3908–3915. [Google Scholar] [CrossRef] [PubMed]
- Hartman, Z.C.; Poage, G.M.; den Hollander, P.; Tsimelzon, A.; Hill, J.; Panupinthu, N.; Zhang, Y.; Mazumdar, A.; Hilsenbeck, S.G.; Mills, G.B.; et al. Growth of triple-negative breast cancer cells relies upon coordinate autocrine expression of the proinflammatory cytokines IL-6 and IL-8. Cancer Res. 2013, 73, 3470–3480. [Google Scholar] [CrossRef] [PubMed]
- Ansari, L.; Shiehzadeh, F.; Taherzadeh, Z.; Nikoofal-Sahlabadi, S.; Momtazi-Borojeni, A.A.; Sahebkar, A.; Eslami, S. The most prevalent side effects of pegylated liposomal doxorubicin monotherapy in women with metastatic breast cancer: A systematic review of clinical trials. Cancer Gene Ther. 2017, 24, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Saupe, S.; Jäger, E.; Schmidt, M.; Kreienberg, R.; Müller, L.; Otremba, B.J.; Waldenmaier, D.; Dorn, J.; Warm, M.; et al. A randomized phase III study evaluating pegylated liposomal doxorubicin versus capecitabine as first-line therapy for metastatic breast cancer: Results of the PELICAN study. Breast Cancer Res. Treat. 2017, 161, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Joerger, M.; Thürlimann, B. Chemotherapy regimens in early breast cancer: Major controversies and future outlook. Expert Rev. Anticancer Ther. 2013, 13, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Fu, Z.; Shi, M.; Xia, K.; Ji, C.; Xu, P.; Lv, M.; Pan, B.; Dai, L.; Xie, H. Systematic analysis of gene expression pattern in has-miR-760 overexpressed resistance of the MCF-7 human breast cancer cell to doxorubicin. Biomed. Pharmacother. 2015, 69, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Xia, K.; Xu, P.; Sun, E.; Ma, J.; Gao, S.; Zhou, Q.; Zhang, M.; Wang, F.; Chen, F.; et al. miRNA expression patterns in chemoresistant breast cancer tissues. Biomed. Pharmacother. 2014, 68, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, M.; Li, Y.; Ye, S.; Ma, J.; Lu, L.; Lv, W.; Chang, G.; Li, X.; Li, Q.; Wang, S.; et al. MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS ONE 2014, 9, e96228. [Google Scholar] [CrossRef] [PubMed]
- Kovalchuk, O.; Filkowski, J.; Meservy, J.; Ilnytskyy, Y.; Tryndyak, V.P.; Chekhun, V.F.; Pogribny, I.P. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol. Cancer Ther. 2008, 7, 2152–2159. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Li, S.; Boohaker, R.J.; Liu, X.; Zhu, Y.; Zhai, L.; Li, H.; Gu, F.; Fan, Y.; Lang, R.; et al. A microRNA Expression Signature In Taxane-anthracycline-Based Neoadjuvant Chemotherapy Response. J. Cancer 2015, 6, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Xue, A.; Chi, Y.; Xue, J.; Wang, W.; Zhao, Z.; Fan, M.; Yang, C.H.; Shao, Z.-M.; Pfeffer, L.M.; et al. Induction of miRNA-181a by genotoxic treatments promotes chemotherapeutic resistance and metastasis in breast cancer. Oncogene 2016, 35, 1302–1313. [Google Scholar] [CrossRef] [PubMed]
- Gasparini, P.; Cascione, L.; Fassan, M.; Lovat, F.; Guler, G.; Balci, S.; Irkkan, C.; Morrison, C.; Croce, C.M.; Shapiro, C.L.; et al. MicroRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget 2014, 5, 1174–1184. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, M.; Ma, F.; Luo, Y.; Cai, R.; Wang, L.; Xu, N.; Xu, B. Circulating miR-19a and miR-205 in serum may predict the sensitivity of luminal A subtype of breast cancer patients to neoadjuvant chemotherapy with epirubicin plus paclitaxel. PLoS ONE 2014, 9, e104870. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Wang, D.; Li, L.; Yang, S.; Chen, X.; Zhou, S.; Zhong, S.; Zhao, J.; Tang, J. miR-222 promotes drug-resistance of breast cancer cells to adriamycin via modulation of PTEN/Akt/FOXO1 pathway. Gene 2017, 596, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Silver, D.P.; Richardson, A.L.; Eklund, A.C.; Wang, Z.C.; Szallasi, Z.; Li, Q.; Juul, N.; Leong, C.-O.; Calogrias, D.; Buraimoh, A.; et al. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J. Clin. Oncol. 2010, 28, 1145–1153. [Google Scholar] [CrossRef] [PubMed]
- Pogribny, I.P.; Filkowski, J.N.; Tryndyak, V.P.; Golubov, A.; Shpyleva, S.I.; Kovalchuk, O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int. J. Cancer 2010, 127, 1785–1794. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Hao, R.; Cai, Y.; Wang, X.; Huang, G. Knockdown of miR-221 promotes the cisplatin-inducing apoptosis by targeting the BIM-Bax/Bak axis in breast cancer. Tumour Biol. 2016, 37, 4509–4515. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Tian, W.; Chen, H.; Jiang, K. miR-944 functions as a novel oncogene and regulates the chemoresistance in breast cancer. Tumour Biol. 2016, 37, 1599–1607. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Wang, S.; Zhao, Y.; Zhang, Z.; Qin, C.; Yang, X. miR-519d impedes cisplatin-resistance in breast cancer stem cells by down-regulating the expression of MCL-1. Oncotarget 2017, 8, 22003–22013. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zhou, S.; Zheng, M.; Deng, X.; Yi, Y.; Huang, T. miR-199a-3p enhances breast cancer cell sensitivity to cisplatin by downregulating TFAM (TFAM). Biomed. Pharmacother. 2017, 88, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Cataldo, A.; Cheung, D.G.; Balsari, A.; Tagliabue, E.; Coppola, V.; Iorio, M.V.; Palmieri, D.; Croce, C.M. miR-302b enhances breast cancer cell sensitivity to cisplatin by regulating E2F1 and the cellular DNA damage response. Oncotarget 2016, 7, 786–797. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Xiao, X.; Dong, L.; Wan, N.; Zhou, Z.; Deng, H.; Zhang, X. miR-218 regulates cisplatin chemosensitivity in breast cancer by targeting BRCA1. Tumour Biol. 2015, 36, 2065–2075. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Peng, J.; Fu, Y.; An, S.; Rezaei, K.; Tabbara, S.; Teal, C.B.; Man, Y.; Brem, R.F.; Fu, S.W. miR-638 mediated regulation of BRCA1 affects DNA repair and sensitivity to UV and cisplatin in triple-negative breast cancer. Breast Cancer Res. 2014, 16, 435. [Google Scholar] [CrossRef] [PubMed]
- Talbot, D.C.; Moiseyenko, V.; Van Belle, S.; O’Reilly, S.M.; Alba Conejo, E.; Ackland, S.; Eisenberg, P.; Melnychuk, D.; Pienkowski, T.; Burger, H.-U.; et al. Randomised, phase II trial comparing oral capecitabine (Xeloda) with paclitaxel in patients with metastatic/advanced breast cancer pretreated with anthracyclines. Br. J. Cancer 2002, 86, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Chen, S.; Zhou, X.; Xie, L.; Chen, A. 5-FU and ixabepilone modify the microRNA expression profiles in MDA-MB-453 triple-negative breast cancer cells. Oncol. Lett. 2014, 7, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; O’Rourke, M.; Winer, E.; Hochster, H.; Chang, A.; Adamkiewicz, B.; White, R.; McGuirt, C. Vinorelbine as first-line chemotherapy for advanced breast cancer in women 60 years of age or older. Ann. Oncol. 1999, 10, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Ma, T.; Zhang, X.; Lv, M.; Chen, L.; Tang, J.; Zhao, J. MicroRNA expression profiling and bioinformatics analysis of dysregulated microRNAs in vinorelbine-resistant breast cancer cells. Gene 2015, 556, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Vale, N.; Ferreira, A.; Fernandes, I.; Alves, C.; Araújo, M.J.; Mateus, N.; Gomes, P. Gemcitabine anti-proliferative activity significantly enhanced upon conjugation with cell-penetrating peptides. Bioorg. Med. Chem. Lett. 2017. [Google Scholar] [CrossRef] [PubMed]
- Earl, H.M.; Vallier, A.-L.; Hiller, L.; Fenwick, N.; Young, J.; Iddawela, M.; Abraham, J.; Hughes-Davies, L.; Gounaris, I.; McAdam, K.; et al. Effects of the addition of gemcitabine, and paclitaxel-first sequencing, in neoadjuvant sequential epirubicin, cyclophosphamide, and paclitaxel for women with high-risk early breast cancer (Neo-tAnGo): An open-label, 2 × 2 factorial randomised phase III trial. Lancet Oncol. 2014, 15, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.-H.; Tao, Z.-H.; Zhang, J.; Li, T.; Ni, C.; Xie, J.; Zhang, J.-F.; Hu, X.-C. miRNA-21 induces epithelial to mesenchymal transition and gemcitabine resistance via the PTEN/AKT pathway in breast cancer. Tumour Biol. 2016, 37, 7245–7254. [Google Scholar] [CrossRef] [PubMed]
| Overexpressed miRNA | Predicted Targets | Pathway Involved | Reference |
|---|---|---|---|
| Paclitaxel | |||
| miR-Lin28 | Let7a | miRNAs processing | [20] |
| miR-125b | Bak1 | Apoptosis | [21] |
| miR-520h | DAPK2 | PI3K/Akt signaling | [22] |
| miR18a | mTOR signaling | [25] | |
| Docetaxel | |||
| miR-129-3p | CP110 | G2/M progression and apoptosis | [31] |
| miR-141 | EIF4E | Apoptosis | [30] |
| miR-3646 | GSK-3β | β-catenin signaling pathway | [32] |
| miR-452 | APC4 | [33] | |
| miR-663 | HSPG2 | [34] | |
| miR-34a | BCL-2 and CCND1 | Apoptosis | [35] |
| miR-222 and miR-29a | PTEN | Apoptosis | [39] |
| Letrozol + anastrozole | |||
| miR-125b and miR-205 | Akt/mTOR pathway | [53] | |
| Fulvestrant | |||
| miR-221 and miR-222 | AXIN2, SFRP2, CHD8 and NLK | p53, TGF-β, MAPK, Notch, ErbB and Jak-STAT | [56] |
| miR-21 | PTEN | PI3K-Akt-mTOR pathway | [57] |
| Tamoxifen | |||
| miR-10b | HDAC4 | Epithelial-mesenchymal transition | [63] |
| miR-210 | RAD52 | Invasion, proliferation and migration | [70] |
| mirR-155 | SOC6 | STAT3 signaling pathway | [62] |
| Trastuzumab | |||
| miR-21 | PTEN | PI3K-Akt-mTOR/epithelial-to-mesenchymal transition and inflammatory signals | [67] |
| miR-150-5p and miR-4734 | [68] | ||
| miR-210 | RAD52 | Cell survival | [69] |
| Doxorrubicin miR-141 miR-200c miR-31 | MAPK signaling pathway, regulation of the actin cytoskeleton, cytokine-cytokine receptor interaction | [90] | |
| miR-155p miR-21-3p miR-181a-5p miR-181b-5p miR-183-5p | PTEN/Akt, MAPK, MDR1, RhoA, FOXO3 and PDCD4 pathway | [91] | |
| miR-125b miR-141 | cell cycle control and apoptotic pathways | [93] | |
| miR-155 miR-493 miR-30e miR-27a | [95] | ||
| miR-181a | Bax | [94] | |
| Epirubicin miR-19a miR-205 | PTEN | [96] | |
| Adriamycin miR-222 | PTEN | PTEN, Akt/FOXP1 pathway | [97] |
| Cisplatin miR-146a miR-10a miR-221/222 miR-345 miR200b and miR-200c | MRP1 | EMT efflux of drugs | [99] |
| miR-221 | BIM-Bax/Bak axis | [100] | |
| miR-944 | BNIP3 | cell proliferation, migration and invasion | [101] |
| Gemcitabine miR-21 | PTEN | EMT | [113] |
| Vinorelbine miR-138-5p, miR-182-5p, miR-18a-5p, miR-193b-3p, miR-199a-5p, miR-210-3p, miR-21-5p, miR-378a-3p, miR-4262, miR-4725-5p and miR-92b-3p | MAPK, mTOR, Wnt, and TGF-β | [110] |
| Overexpressed miRNA | Predicted Targets | Pathway Involved | Reference |
|---|---|---|---|
| Paclitaxel | |||
| miR-451 | Bcl-2 | Apoptosis | [23] |
| miR-100 | mTOR | Cell proliferation and survival | [24] |
| Docetaxel | |||
| miR-139-5p | Notch1 | Cell growth and apoptosis | [36] |
| miR-205 | cell proliferation and clonogenic potential | [37] | |
| miR-125a-3p | BRCA1 | [38] | |
| Fulvestran | |||
| miR-214 | UCP2 | Autophagy | [55] |
| Tamoxifen | |||
| miR-30c | HER2 and RAC1 signaling pathway | [59] | |
| MiR-10a and miR-126 | [60] | ||
| Lapatinib | |||
| miR-16 | CCNJ and FUBP1 | PI3K/Akt signaling pathways | [80] |
| Cisplatin miR-519d | MCL-1 | anti-apoptotic signaling pathway | [102] |
| miR-199a-3p | TFAM | mitochondrial biogenesis | [103] |
| miR-302b | E2F1 | E2f1-ATM axis | [104] |
| miR-218, miR-638 | BRCA1 | DNA reparation, cell proliferation and invasion | [105,106] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos-Parra, A.D.; Mitznahuatl, G.C.; Pedroza-Torres, A.; Romo, R.V.; Reyes, F.I.P.; López-Urrutia, E.; Pérez-Plasencia, C. Micro-RNAs as Potential Predictors of Response to Breast Cancer Systemic Therapy: Future Clinical Implications. Int. J. Mol. Sci. 2017, 18, 1182. https://doi.org/10.3390/ijms18061182
Campos-Parra AD, Mitznahuatl GC, Pedroza-Torres A, Romo RV, Reyes FIP, López-Urrutia E, Pérez-Plasencia C. Micro-RNAs as Potential Predictors of Response to Breast Cancer Systemic Therapy: Future Clinical Implications. International Journal of Molecular Sciences. 2017; 18(6):1182. https://doi.org/10.3390/ijms18061182
Chicago/Turabian StyleCampos-Parra, Alma D., Gerardo Cuamani Mitznahuatl, Abraham Pedroza-Torres, Rafael Vázquez Romo, Fany Iris Porras Reyes, Eduardo López-Urrutia, and Carlos Pérez-Plasencia. 2017. "Micro-RNAs as Potential Predictors of Response to Breast Cancer Systemic Therapy: Future Clinical Implications" International Journal of Molecular Sciences 18, no. 6: 1182. https://doi.org/10.3390/ijms18061182