2.3. Functional Annotations
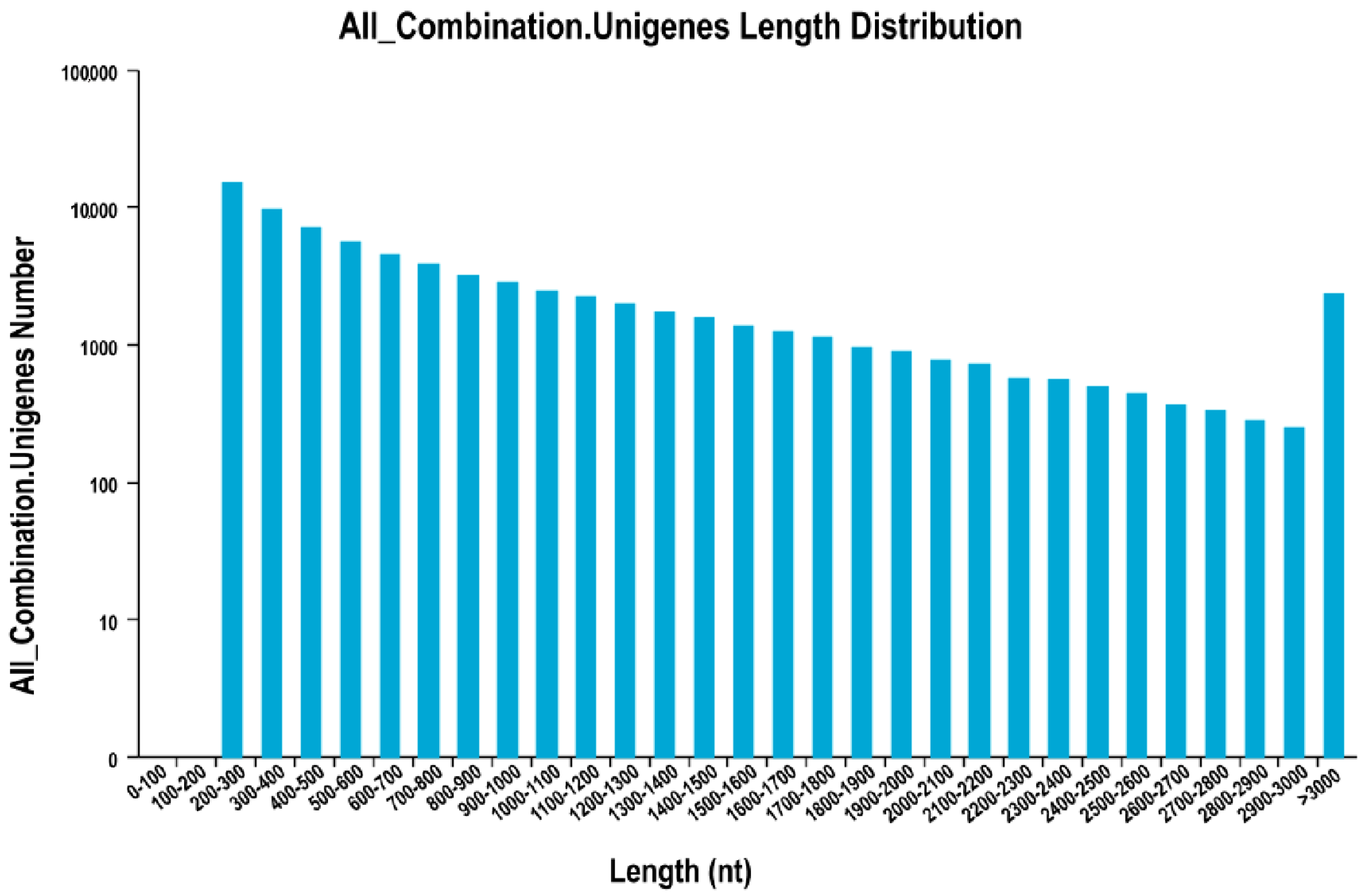
All unigenes generated by HiSeq 2500 were aligned to information in the non-redundant (NR), Clusters of Orthologous Groups (COG), euKaryotic Orthologous Groups (KOG), Gene Ontology (GO), Swiss-Prot, Protein family (Pfam), and Kyoto Encyclopedia of Genes and Genomes (KEGG) protein databases by BLASTx and BLASTn. After all of the amino acid sequences were predicted, we compared them with the Pfam database to obtain annotation information. In total, 35,793 unigenes (48.28%) were aligned to homologous sequences in those public databases. Of them, 14,618 (19.72%) were 300 to 1000 bp long, while 17,140 (23.12%) were longer than 1000 bp (
Table 3).
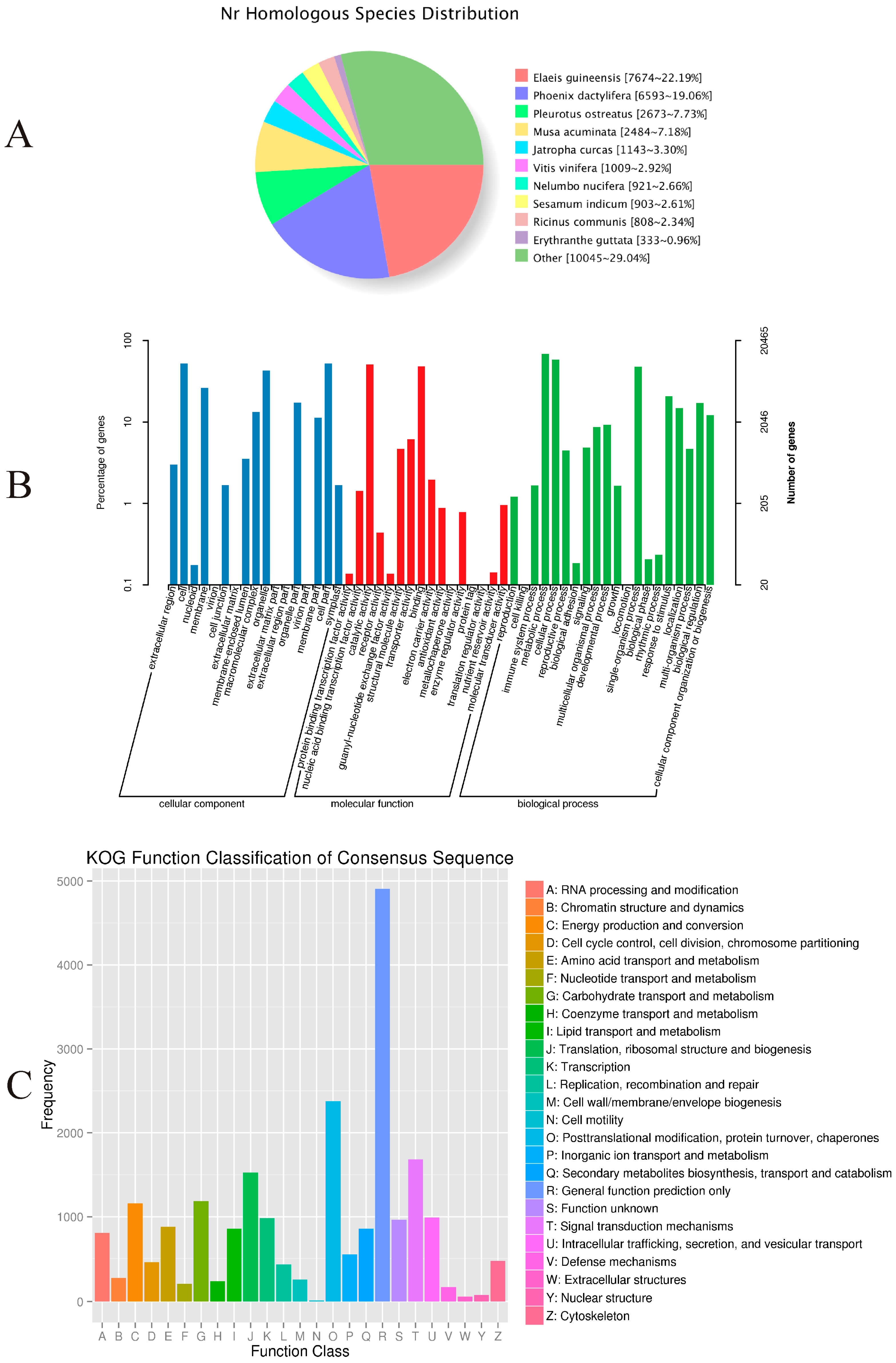
Approximately 46.68% (34,601) of the unigenes were annotated with reference to the NR database. As shown in
Figure 3A, 22.19% had top matches to the sequences from
Elaeis guineensis, 19.06% to the sequences from
Phoenix dactylifera, and 7.73% to the sequences from
Pleurotus ostreatus.
Among the 34,601 most significant BLASTx hits against the NR database, 20,465 unigenes could be assigned to one or more GO terms. These were assigned to 53 functional groups (
Figure 3B). Among those categorized in “cellular components”, 10,797 (52.76%) were related to “cell part” (GO: 0044464), followed by “membrane” (GO: 0016020; 5395, or 26.36%). Within the molecular functions, the assignments were mostly enriched in “catalytic activity” (GO: 0003824; 10,444) and “binding” (GO: 0005488; 9905). The GO terms of biological processes were mainly grouped into “metabolic process” (GO: 0008152; 14,065) and “cellular process” (GO: 0009987; 11,945).
The COG assignments were used for further evaluation of the completeness of our
P. sibiricum transcriptome library and the reliability of the annotation process. Overall, 12,175 (16.42%) unigenes were assigned to 25 COG categories (
Figure 3C). Among these, the most numerous were unigenes belonging to the “General function prediction only” (2965, or 24.35% of the total), followed by “Replication, recombination, and repair” (1441, 11.83%), and “Translation, ribosomal structure, and biogenesis” (1426, 11.74%). Another 946 unigenes (7.77%) were assigned to “carbohydrate transport and metabolism”.
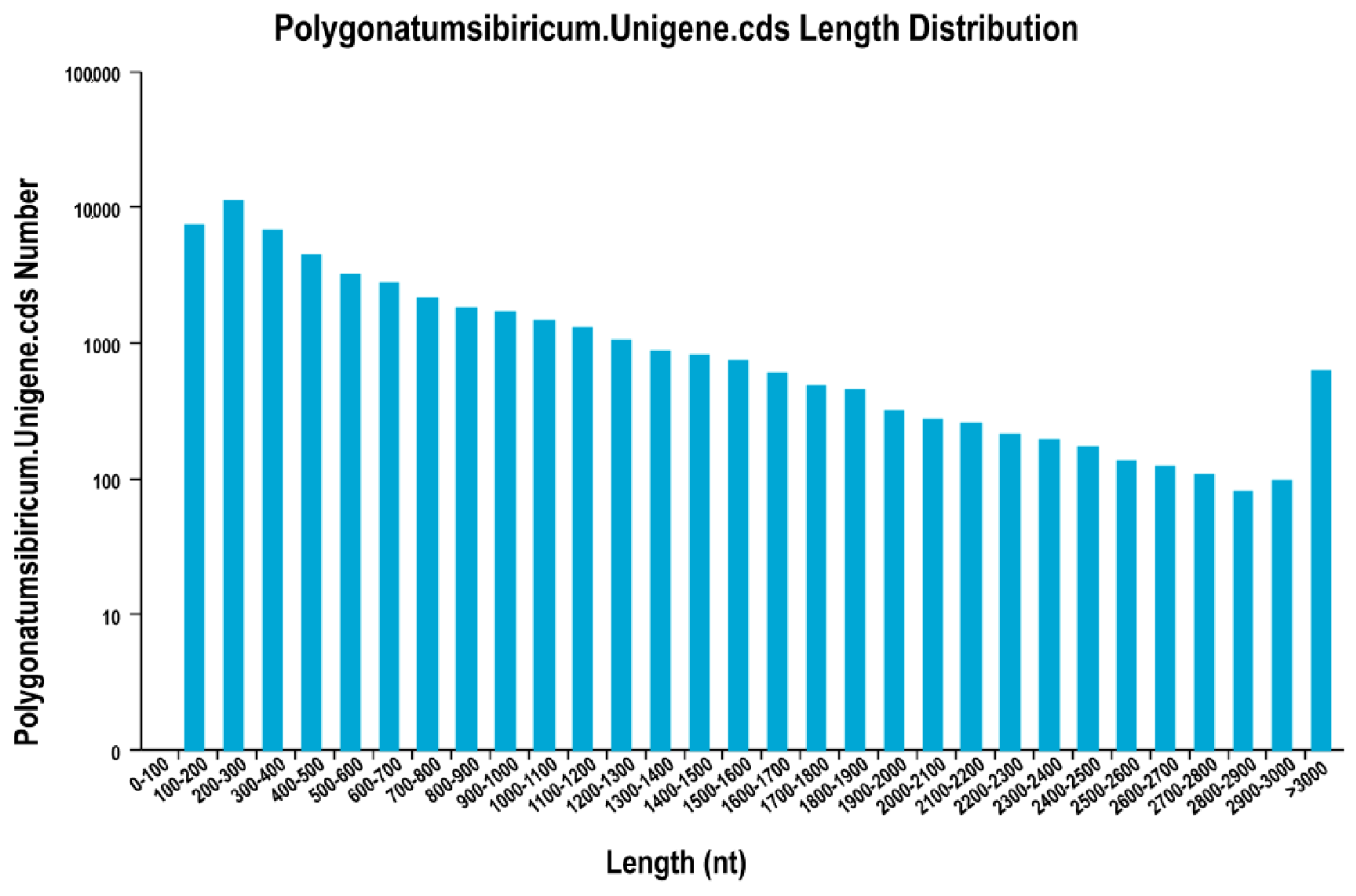
Using the three public protein databases, we obtained 51,461 coding sequences (CDSs), which accounted for 69.42% of all unigenes identified here. The length distribution of CDSs (
Figure 4) indicated that 33,862 were 200 to 1000 bp long, and that 34.20% were longer than 500 bp. Another 8014 (15.57%) were 1000 to 2000 bp long, 1636 (3.19%) were 2000 to 3000 bp long, and 629 (1.22%) were longer than 3000 bp.
2.5. Candidate Genes Involved in PSP Biosynthesis
To understand the biosynthesis of PSP and to identify the responsible genes, we annotated the transcripts related to the KO00500 and KO00520 pathway. Mainly based on the KEGG database, we determined the key enzyme genes involved in these pathways (
Table 6).
Plant polysaccharides are formed by the active nucleotide-diphospho-sugar (NDP-sugar) precursors, which are added to the residues of polysaccharides and glycoconjugates by the action of various glycosyltransferases (GTs) [
12,
13]. According to previous studies [
12,
13,
14,
15,
16,
17,
18,
19,
20], PSP biosynthesis can be divided into three main stages. Firstly, sucrose is converted to Glc-1P and Fru-6P. During these processes, many enzymes play their roles, such as the β-fructofuranosidase (encoded by
sacA) [
14] that converts sucrose to Glc-6P and Fructose, and Phosphoglucomutase (encoded by
pgm) that isomerizes Glc-6P to Glc-1P [
15]. Hexokinase (encoded by
HK) [
16] and Fructokinase (encoded by
scrK) [
17] also take part in the biosynthesis of Fru-6P. Secondly, uridine diphosphate glucose (UDP-Glc) is derived from Glc-1P immediately [
18], and Fru-6P is converted to GDP-Man indirectly [
19]. Based on UDP-Glc and guanosine diphospho mannose (GDP-Man), other NDP sugars are further converted through the action of NDP-sugar interconversion enzymes (NSEs) [
20], such as UDP-glucose 4-epimerase (
GALE), UDP-
d-galactose dehydrogenase (
UGD), UDP-glucuronate 4-epimerase (
UGE), UDP-glucose 6-dehydrogenase (
UGDH), UDP-arabinose 4-epimerase (
UXE), UDP-glucose 4,6-dehydratase (
RHM), 3,5-epimerase-4-reductase (
UER1), and GDP-mannose 4,6-dehydratase (
GMDS). Finally, various NDP-sugars form growing polysaccharide chains by the action of glycosyltransferases (GTs).
In plants, UDP-Glc is a key precursor of nucleoside diphosphate (NDP)-sugars, and it is derived from 1-P-glucose (Glu-1-P) by the action of Uridine-diphosphate glucose pyrophosphorylase (encoded by
galU) [
18,
21]. In the
P. sibiricum library, Unigene 124,676 was identified as a homologue of
galU. This 1153 bp unigene contained a complete open reading frame, and shared the highest amino acid identity (86%) with
galU from
Ricinus communis (Accession Number: XM_002526548.2) (
Figures S1 and S2). Sugars produced by plants, such as the hexose sugars glucose and fructose, serve as the basic material for the production and accumulation of organic compounds in plants. The first step of hexose sugars’ metabolism is to phosphorylate them. Currently, only two kinds of plant enzymes, hexokinases (HKs) and fructokinases (scrKs), have been found to catalyze the conversion of glucose and fructose. Here, we found that the 2017 bp Unigene 137,389 was a
HK homologue that shared the highest amino acid identity (72%) with
HK from
Phoenix dactylifera (Accession Number: XM_008797620.2) (
Figures S1 and S3). Furthermore, the 1803 bp Unigene 107,236 was a homologue of
scrK from
Elaeis guineensis (Accession number: XM_010934340.2) (
Figures S1 and S4).
An examination of the monosaccharide composition (
Table S2) showed that PSPs are formed by the polymerization of NDP-sugars such as UDP-Glc, uridine diphosphate galactose (UDP-Gal), uridine diphosphate arabinose (UDP-Ara), uridine diphosphate rhamnose (UDP-Rha), GDP-Man, and guanosine diphosphate fucose (GDP-Fuc). UDP-Glc is derived from Glc-1P directly, and GDP-Man from Fru-6P indirectly, and the others are derived from UDP-Glc and GDP-Man through the actions of NSEs [
20]. We identified eight subclasses of NSEs in
P. sibiricum, including
GALE,
UGD,
UGE,
UGDH,
UXE,
RHM,
UER1, and
GMDS. In all, 47 transcripts encoding NSEs were found in this species (
Table 6,
Table S3). By comparison of the fragments per kilobase of transcript per Million mapped reads (FPKM) from different RNA-Seq libraries, the most abundant transcript was for mannose-1-phosphate guanylyltransferase (
GMPP), followed by
pgm (phosphoglucomutase) and
sacA (β-fructofuranosidase).
Protein encoded by pgm is responsible for the production of Glc-1P, using Glc-6P as a precursor. The enzyme encoded by UGDH converts UDP-Glu into UDP-GluA, while the enzyme encoded by GMPP converts Man-1P into GDP-Man. SacA converts sucrose into fructose and Glc-6P. The relatively higher abundance of such transcripts determined here suggests that the biological process of conversion from Glc-6P to Glc-1P should be very active.
2.6. Analysis of Differential Gene Expression in Lueyang, Shaanxi (SXLY) and Luoyang, Henan (HNLY) Germplasms
We used DESeq to analyze differences in gene expression between SXLY and HNLY. In the screening process, the false discovery rate (FDR) was set at <0.01 and the fold change (FC) was >2. The Volcano Plot presented a clear visual of the relationship between the FDR and FC for all genes of interest, enabling us to view expression levels quickly. As shown in
Figure 5A, 435 genes were upregulated and 494 were downregulated. These upregulated and downregulated (929) genes were assigned to 50 functional groups (
Figure 5B). Among those categorized in “biological processes”, 132 were related to “metabolic process” (GO: 0008152), followed by “cellular process” (GO: 0009987; 108). Within the molecular functions, the assignments were mostly enriched in “catalytic activity” (GO: 0003824; 146) and “binding” (GO: 0005488; 112). The GO terms of cellular components were mainly grouped into “cell part” (GO: 0044464; 88) and “organelle” (GO: 0043231; 60).
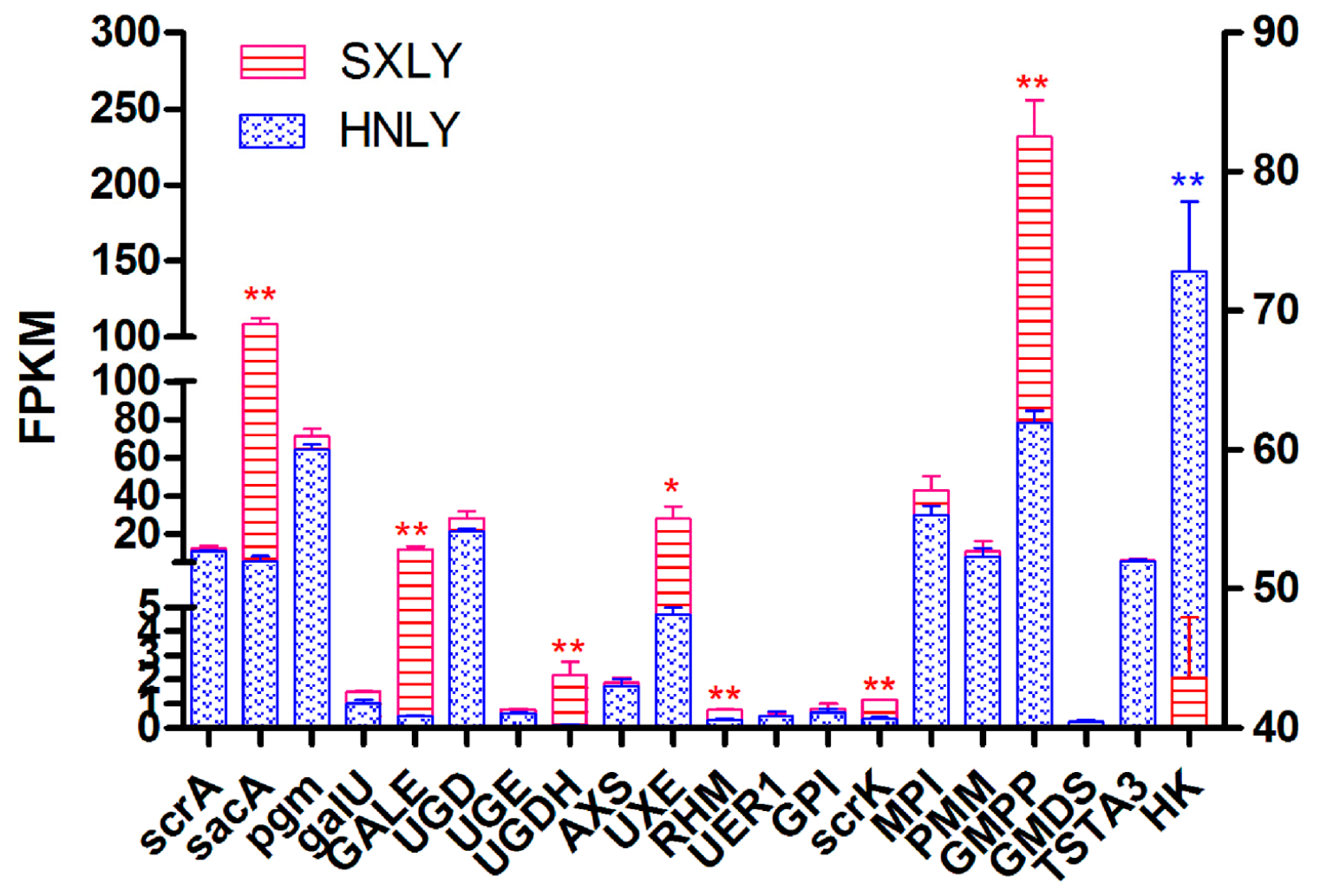
Among these upregulated and downregulated genes, we focused more on those involved in the pathway of PSP biosynthesis. Based on the abundance of their transcripts, as determined through RNA-seq, we selected eight candidate unigenes:
sacA,
GALE,
UGDH,
UXE,
RHM,
scrK,
GMPP, and
HK (
Figure 6). Those sharing the least identity with their homologs in the dataset were
scrK (Unigene 107,236; 53% identity) and
UGDH (Unigene 153,987; 64% identity). Percent identities for the others—
sacA (Unigene 157,362),
GALE (Unigene 51,042),
UXE (Unigene 153,844),
RHM (Unigene 142,186),
HK (Unigene 137,389), and
GMPP (Unigene 151,473)—ranged from 72% to 99%. Details for all of these alignments are shown in
Table S4. This analysis primarily confirmed that our chosen potential unigenes are rather conservative and responsible for PSP biosynthesis.
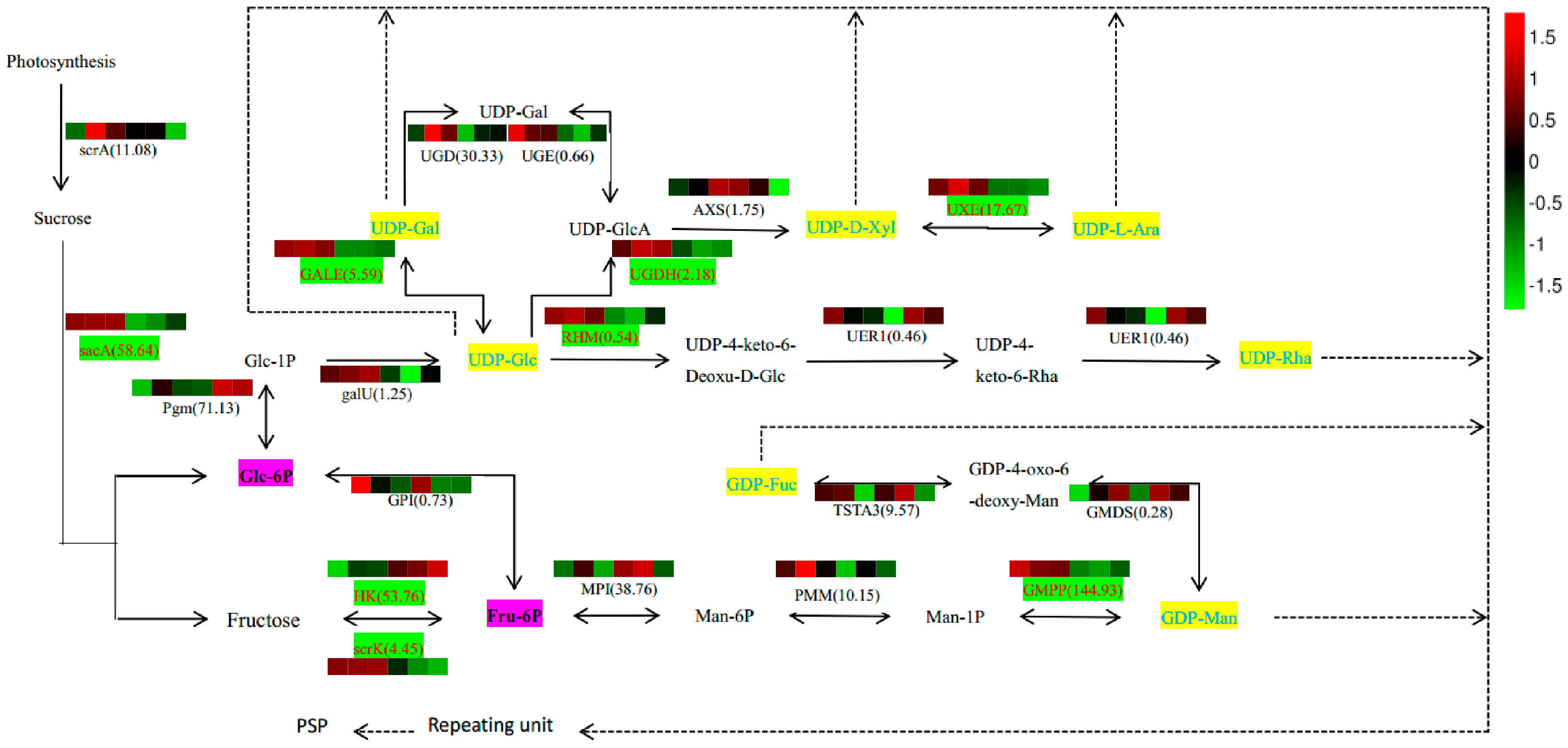
2.7. Analysis of PSP Biosynthetic Pathway
Based on our results that identified constituents in the polysaccharide biosynthesis pathway and monosaccharide composition (
Table S2), we outlined potential biosynthetic pathways for PSP formation from sucrose (
Figure 7). In this scheme, the biosynthesis of PSP comprises three main steps. First, Glc-1P is converted to UDP-Glc immediately, and Fru-6P to GDP-Man indirectly, and then converted in the second step into other NDP-sugars by NSEs. Finally, the NDP-sugars are added to the polysaccharide by several GTs. Various color blocks represent the logarithms of FPKM values for different samples, using normalized data processing and plotting on a heatmap produced with R software. The first three colors in that figure are for SXLY samples; the latter three, for HNLY samples. The key enzymes are highlighted in red and are responsible for the biosynthesis of Glc-6P (
sacA), Fru-6P (
scrK), UDP-Gal (
GALE), UDP-GlcA (
UGDH), UDP-Ara (
UXE), UDP-4-keto-6-Deoxu-D-Glc (
RHM), fructose (
HK), and GDP-Man (
GMPP). Whereas expression of
sacA,
scrK,
GALE,
UGDH,
RHM,
GMPP, and
UXE are positively correlated to the PSP content, that of
HK is negatively correlated.
Various NDP-sugars form a growing polysaccharide chain by the action of glycosyltransferases (GTs) (EC: 2.4.x.y). In the
P. sibiricum library, we identified 380 unigenes encoding GTs, based on KGG annotations. Among them, 18 unigenes are UDP glycosyltransferases (UGTs) (
Table S5). In general, approximately 48% of UGTs have a plant secondary product glycosyltransferase (PSPG) box [
22]. This motif contains 44 conserved amino acids. We used WebLogo (
http://weblogo.berkeley.edu/logo.cgi) to examine the typical PSPG amino acid sequences for these 18 UGTs. As shown in
Figure 8, the font size represents the intensity of the conserved properties for amino acid residues. That is, the 1st and 22nd tryptophan (W); 3rd, 34th, and 39th proline (P); 4th and 44th glutamine (Q); 8th leucine (L); 14th, 21st, and 32nd glycine (G); and 19th histidine (H) are more conserved than any of the other amino acids in the motif. A previous study of the structure-activity relationship in UGTs has shown that the N-terminal domain is the acceptor binding site and the C-terminal domain is the donor response site [
23].
2.8. Real-Time PCR Analysis
Performing a qRT-PCR analysis enabled us to determine the abundance of transcripts of genes for enzymes related to PSP biosynthesis. After selecting the reference genes in
P. sibiricum (
Table S6), we chose
β-Tubulin (
TUB) as our internal reference. We then screened eight genes as candidate unigenes based on the significantly broad differences in their levels of expression between germplasms (
Figure 6). Among these candidate unigenes, we predicted that those encoding
sacA,
scrK, and
HK should function upstream of the proposed pathway for PSP biosynthesis (
Figure 7), while those encoding
GALE,
RHM,
UGDH,
UXE, and
GMPP should act downstream.
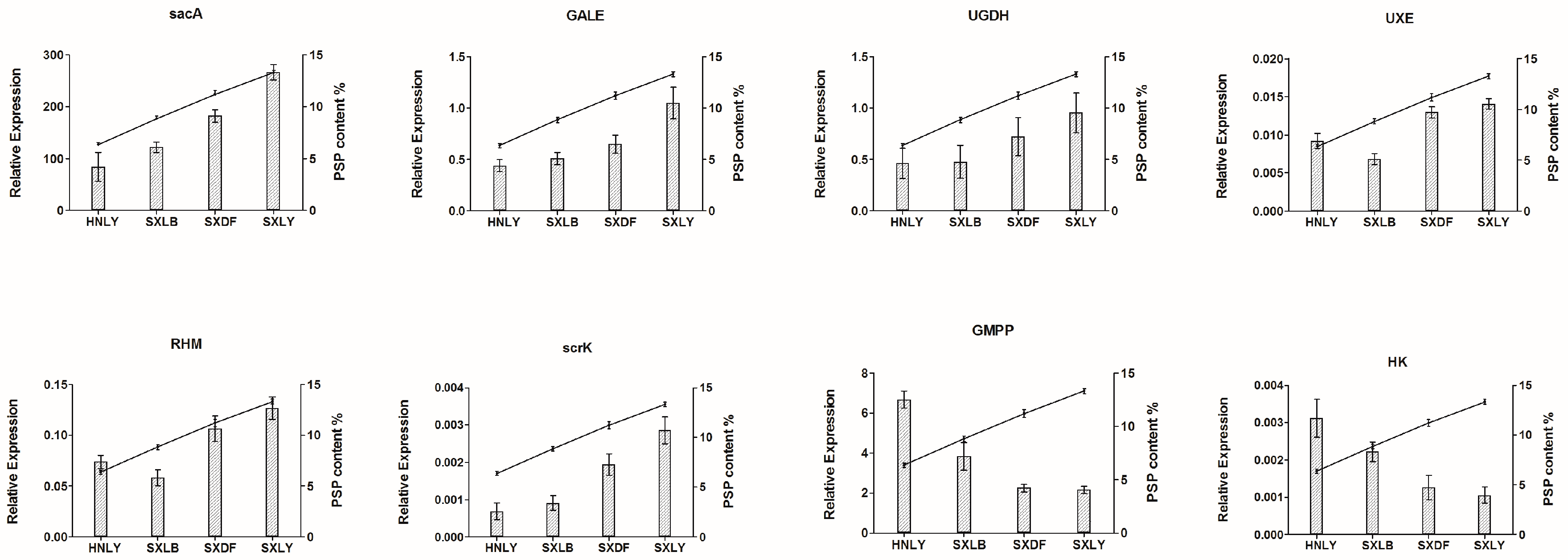
We also examined the relative expression of these eight candidate genes in four germplasms: SXLY, HNLY, SXLB, and SXDF (
Figure 9). That of
sacA,
GALE,
UGDH,
UXE,
RHM, and
scrK was positively correlated with polysaccharide content, while the relative expression of
GMPP and
HK was negatively correlated. By comparing the results of qRT-PCR and RNA-seq, we found that only the expression pattern of
GMPP was inconsistent between the two results. The enzymes encoded by
GMPP can convert Man-1P into GDP-Man in both directions. Maybe too much accumulation of PSP can cause
GMPP negative feedback regulation, leading to the opposite results between the RNA-seq and qRT-PCR.