Characterization of a Potential Probiotic Lactobacillus brevis RK03 and Efficient Production of γ-Aminobutyric Acid in Batch Fermentation
Abstract
:1. Introduction
2. Results
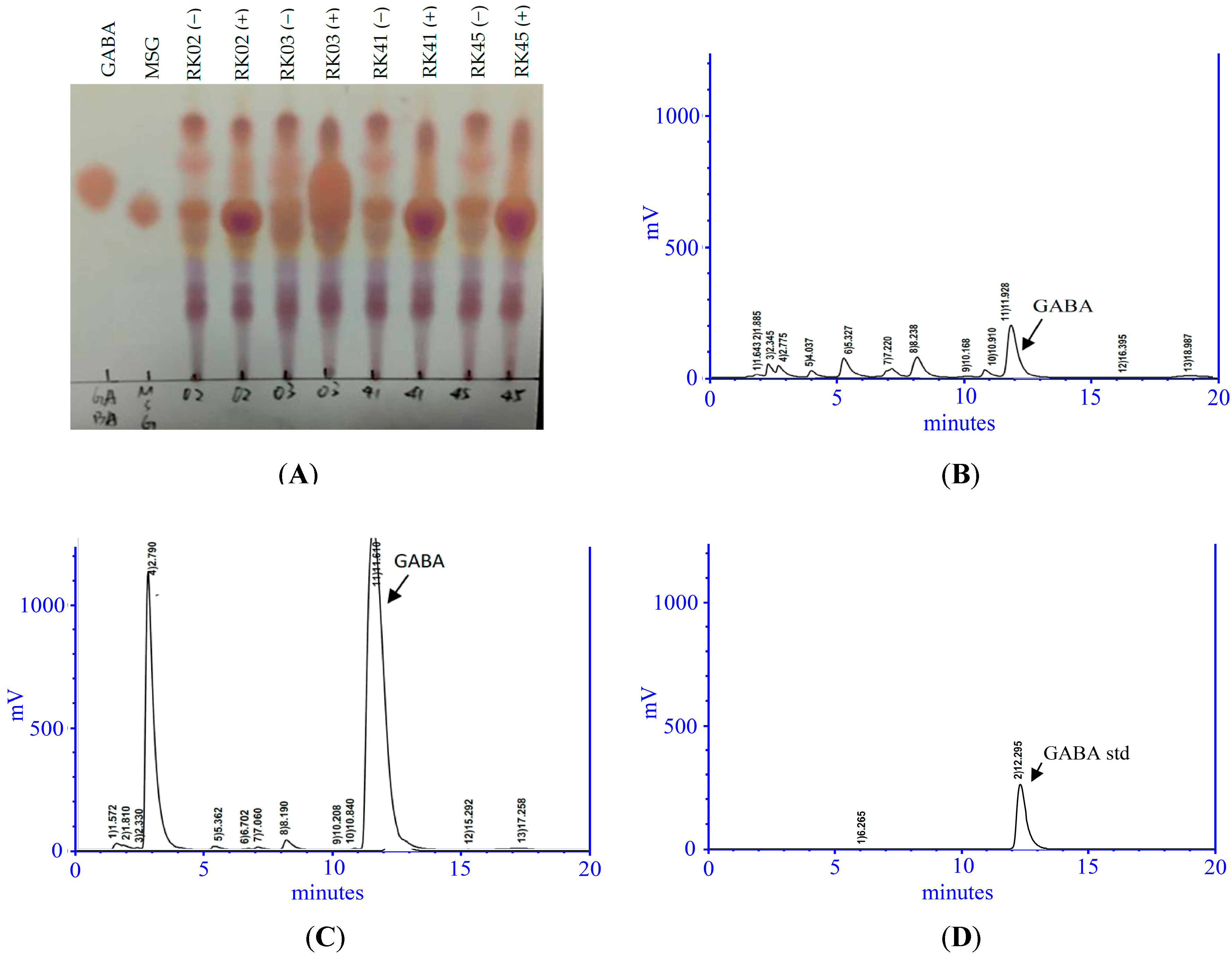
2.1. Isolation of a High-GABA-Producing LAB from Saltwater Fish
2.2. Effect of Initial Glutamic Acid Concentrations on Growth Profile and GABA Production
2.3. Effect of Initial pH and Temperature on Growth Profile and GABA Production
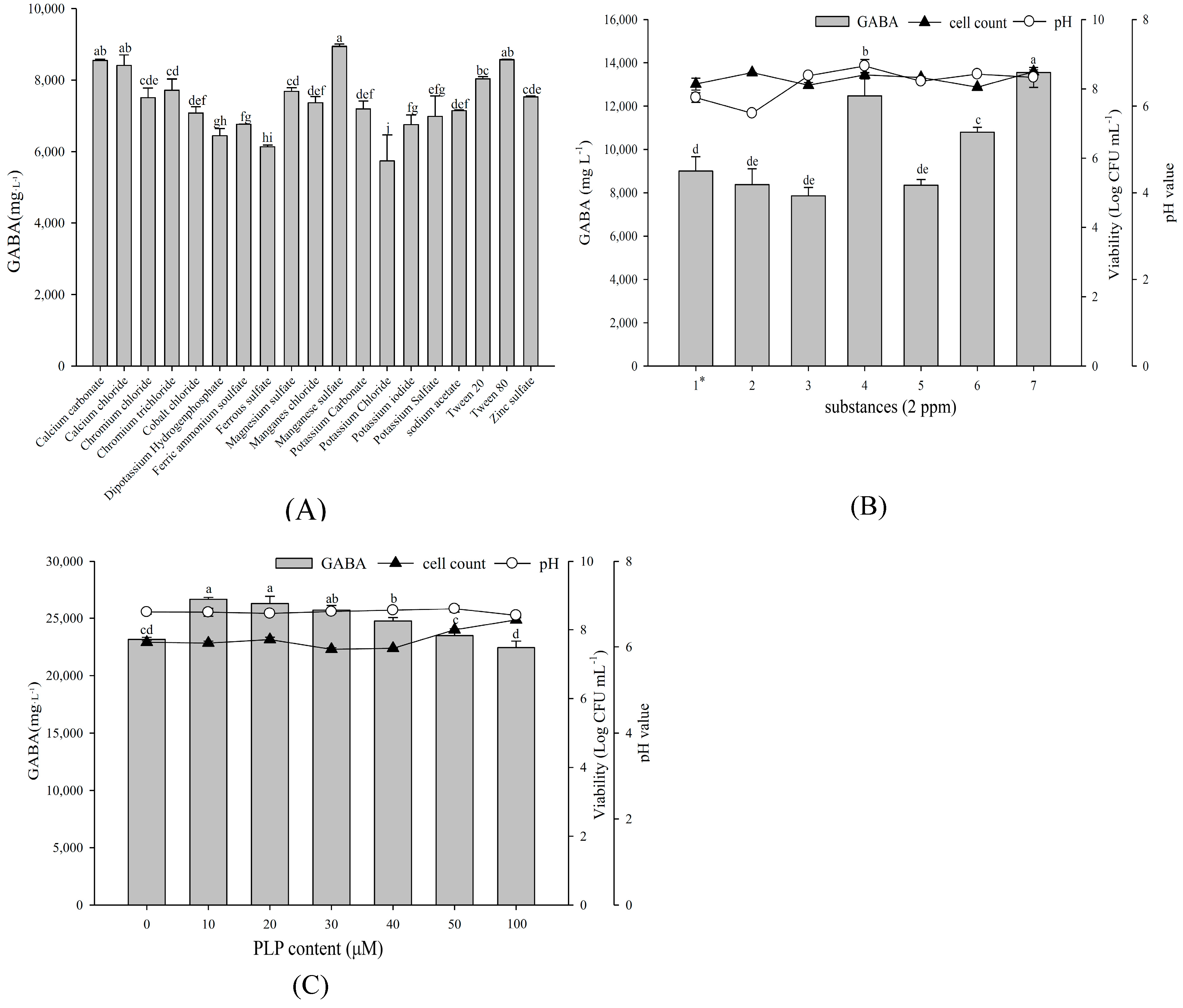
2.4. Effect of Organic Carbon and Nitrogen Sources on Growth Profile and GABA Production
2.5. Effect of Various Substances on Growth Profile and GABA Production
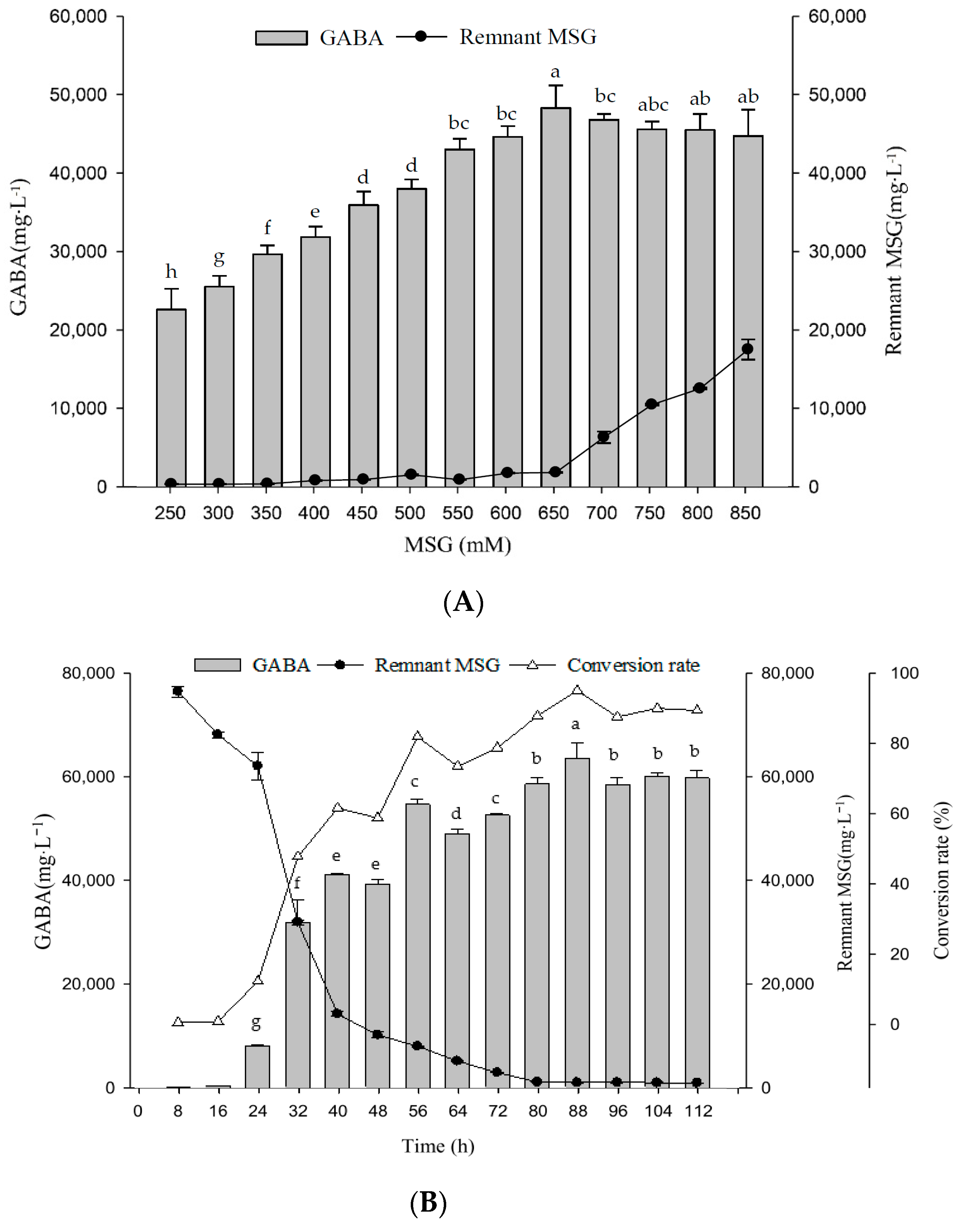
2.6. Optimal MSG Concentrations in G Broth on Growth Profile and GABA Production
2.7. Acid, Intestinal, Gastrointestinal Juice Tolerances and Antibiotic Susceptibilities
3. Discussion
4. Materials and Methods
4.1. Isolation and Identification of GABA-Producing LAB
4.1.1. Isolation of GABA-Producing LAB from Fish Intestines
4.1.2. Gram Staining, Catalase, Oxidase, and Acid Production Tests
4.1.3. Amplification and Sequencing of 16S rDNA
4.1.4. Preservation and Culture of LAB Isolates
4.2. Measurement of GABA Content
4.2.1. Thin Layer Chromatography (TLC) Assay
4.2.2. High-Performance Liquid Chromatography (HPLC) Analysis
4.3. Single Parameter Optimization
4.4. Culture Recipes for Larger Scale GABA Production
4.4.1. Organic Carbon Sources
4.4.2. Nitrogen Sources
4.4.3. Growth Factors
4.4.4. Pyridoxal Phosphate (PLP) Compound
4.4.5. Monosodium Glutamate (MSG) Concentrations
4.5. In Vitro Test of Gastric Acidity: Tolerance to Simulated Intestinal Juice and Gastrointestinal Juice
4.6. Resistance of L. brevis RK03 to Antibiotics
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Narayan, V.S.; Nair, P. Metabolism, enzymology and possible roles of 4-aminobutyrate in higher plants. Phytochemistry 1990, 29, 367–375. [Google Scholar] [CrossRef]
- Zhang, H.; Yao, H.-Y.; Chen, F.; Wang, X. Purification and characterization of glutamate decarboxylase from rice germ. Food Chem. 2007, 101, 1670–1676. [Google Scholar] [CrossRef]
- Ko, C.Y.; Lin, H.-T.V.; Tsai, G.J. Gamma-aminobutyric acid production in black soybean milk by Lactobacillus brevis FPA 3709 and the antidepressant effect of the fermented product on a forced swimming rat model. Process Biochem. 2013, 48, 559–568. [Google Scholar] [CrossRef]
- Nishimura, M.; Yoshida, S.-I.; Haramoto, M.; Mizuno, H.; Fukuda, T.; Kagami-Katsuyama, H.; Tanaka, A.; Ohkawara, T.; Sato, Y.; Nishihira, J. Effects of white rice containing enriched gamma-aminobutyric acid on blood pressure. J. Tradit. Complement. Med. 2016, 6, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Taneera, J.; Jin, Z.; Jin, Y.; Muhammed, S.J.; Zhang, E.; Lang, S.; Salehi, A.; Korsgren, O.; Renström, E.; Groop, L. γ-Aminobutyric acid (GABA) signalling in human pancreatic islets is altered in type 2 diabetes. Diabetologia 2012, 55, 1985–1994. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhao, H.; Zhang, C.; Lu, Y.; Zhu, X.; Lu, Z. γ-Aminobutyric acid-rich yogurt fermented by Streptococcus salivarius subsp. thermophiles fmb5 apprars to have anti-diabetic effect on streptozotocin-induced diabetic mice. J. Funct. Foods 2016, 20, 267–275. [Google Scholar] [CrossRef]
- Marques, T.M.; Patterson, E.; Wall, R.; O’Sullivan, O.; Fitzgerald, G.; Cotter, P.D.; Dinan, T.; Cryan, J.; Ross, R.P.; Stanton, C. Influence of GABA and GABA-producing Lactobacillus brevis DPC 6108 on the development of diabetes in a streptozotocin rat model. Benef. Microbes 2016, 7, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-Y.; Ho, K.-J.; Hsieh, Y.-J.; Wang, L.-T.; Mau, J.-L. Contents of lovastatin, γ-aminobutyric acid and ergothioneine in mushroom fruiting bodies and mycelia. LWT Food Sci. Technol. 2012, 47, 274–278. [Google Scholar] [CrossRef]
- Song, N.E.; Baik, S.H. Identification and characterization of high GABA and low biogenic amine producing indigenous yeasts isolated from Korean traditional fermented Bokbunja (Rubus coreanus Miquel) wine. J. Biotechnol. 2014. [Google Scholar] [CrossRef]
- Franciosi, E.; Carafa, I.; Nardin, T.; Schiavon, S.; Poznanski, E.; Cavazza, A.; Larcher, R.; Tuohy, K.M. Biodiversity and γ-aminobutyric acid production by lactic acid bacteria isolated from traditional Alpine raw cow’s milk cheeses. Biomed. Res. Int. 2015. [Google Scholar] [CrossRef] [PubMed]
- Ramos, C.L.; Thorsen, L.; Schwan, R.F.; Jespersen, L. Strain-specific probiotics properties of Lactobacillus fermentum, Lactobacillus plantarum and Lactobacillus brevis isolates from Brazilian food products. Food Microbiol. 2013, 36, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Sanchart, C.; Rattanaporn, O.; Haltrich, D.; Phukpattaranont, P.; Maneerat, S. Technological and safety properties of newly isolated GABA-producing Lactobacillus futsaii strains. J. Appl. Microbiol. 2016, 121, 734–745. [Google Scholar] [CrossRef] [PubMed]
- Binh, T.T.T.; Ju, W.-T.; Jung, W.-J.; Park, R.-D. Optimization of γ-amino butyric acid production in a newly isolated Lactobacillus brevis. Biotechnol. Lett. 2014, 36, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Qiu, T.; Gao, D.; Cao, Y. Medium optimization for production of gamma-aminobutyric acid by Lactobacillus brevis NCL912. Amino Acids 2010, 38, 1439–1445. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Qiu, T.; Huang, G.; Cao, Y. Production of gamma-aminobutyric acid by Lactobacillus brevis NCL912 using fed-batch fermentation. Microb. Cell Fact. 2010, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Villegas, J.M.; Brown, L.; de Giori, G.S.; Hebert, E.M. Optimization of batch culture conditions for GABA production by Lactobacillus brevis CRL 1942, isolated from quinoa sourdough. LWT Food Sci. Technol. 2016, 67, 22–26. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, L.; Gao, Q.; Yu, S.M.; Li, L.; Gao, N.F. The two-step biotransformation of monosodium glutamate to GABA by Lactobacillus brevis growing and resting cells. Appl. Microbiol. Biotechnol. 2012, 94, 1619–1627. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, Y.-H.; Liaw, W.-C.; Kuo, J.-M.; Deng, C.-S.; Wu, C.-H. Hydrogel film-immobilized Lactobacillus brevis RK03 for γ-aminobutyric acid production. Int. J. Mol. Sci. 2017, 18, 2324. [Google Scholar] [CrossRef] [PubMed]
- Gobbetti, M.; Ferranti, P.; Smacchi, E.; Goffredi, F.; Addeo, F. Production of angiotensin-I-convertin-enzyme-inhibitory peptides in fermented milks started by Lactobacillus delbrueckiisubsp. bulgaricus SS1 and Lactococcus lactissubsp. cremoris FT4. Appl. Environ. Microb. 2000, 66, 3898–3904. [Google Scholar] [CrossRef]
- Nomura, M.; Kimoto, H.; Someya, Y.; Furukawa, S.; Suzuki, I. Production of γ-aminobutyric acid by cheese starters during cheese ripening. J. Dairy Sci. 1998, 81, 1486–1491. [Google Scholar] [CrossRef]
- Komatsuzaki, N.; Shima, J.; Kawamoto, S.; Momose, H.; Kimura, T. Production of γ-aminobutyric acid (GABA) by Lactobacillus paracasei isolated from traditional fermented foods. Food Microbiol. 2005, 22, 497–504. [Google Scholar] [CrossRef]
- Ferrando, V.; Quiberoni, A.; Reinheimer, J.; Suárez, V. Functional properties of Lactobacillus plantarum strains: A study in vitro of heat stress influence. Food Microbiol. 2016, 54, 154–161. [Google Scholar] [CrossRef]
- Hiraga, K.; Ueno, Y.; Sukontasing, S.; Tanasupawat, S.; Oda, K. Lactobacillus senmaizukei sp. nov., isolated from Japanese pickle. Int. J. Syst. Evol. Microbiol. 2008, 58, 1625–1629. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-Y.; Lü, F.-X.; Lu, Z.-X.; Bie, X.-M.; Jiao, Y.; Sun, L.-J.; Yu, B. Production of γ-aminobutyric acid by Streptococcus salivarius subsp. thermophilus Y2 under submerged fermentation. Amino Acids 2008, 34, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Lee, M.Y.; Ji, G.E.; Lee, Y.S.; Hwang, K.T. Production of γ-aminobutyric acid in black raspberry juice during fermentation by Lactobacillus brevis GABA100. Int. J. Food Microbiol. 2009, 130, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Gao, D.; Cao, Y.; Xu, H. A high γ-aminobutyric acid-producing Lactobacillus brevis isolated from Chinese traditionalpaocai. Ann. Microbiol. 2008, 58, 649–653. [Google Scholar] [CrossRef]
- Ueno, Y.; Hayakawa, K.; Takahashi, S.; Oda, K. Purification and characterization of glutamate decarboxylase from Lactobacillus brevis IFO 12005. Biosci. Biotechnol. Biochem. 1997, 61, 1168–1171. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.-J.; Nam, Y.-D.; Lee, S.-Y.; Park, S.-L.; YI, S.-H.; Lim, S.-I. Expression and characterization of a glutamate decarboxylase from Lactobacillus brevis 877G producing γ-aminobutyric acid. Biosci. Biotechnol. Biochem. 2013, 77, 853–856. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; Ladero, V.; Alvarez, M.A.; Lucas, P.M. Putrescine production via the ornithine decarboxylation pathway improves the acid stress survival of Lactobacillus brevis and is part of a horizontally transferred acid resistance locus. Int. J. Food Microbiol. 2014, 175, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; de Palencia, P.F.; Romano, A.; Fernández, M.; Lucas, P.; Spano, G.; López, P. Biogenic amine production by the wine Lactobacillus brevis IOEB 9809 in systems that partially mimic the gastrointestinal tract stress. BMC Microbiol. 2012, 12, 247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, Y.-H.; Kung, H.-F.; Lin, Q.-L.; Hwang, J.-H.; Cheng, S.-H.; Wei, C.-I.; Hwang, D.-F. Occurrence of histamine and histamine-forming bacteria in kimchi products in Taiwan. Food Chem. 2005, 90, 635–641. [Google Scholar] [CrossRef]
- Son, S.-H.; Jeon, H.-L.; Yang, S.-J.; Lee, N.-K.; Paik, H.-D. In vitro characterization of Lactobacillus brevis KU15006, an isolate from kimchi, reveals anti-adhesion activity against foodborne pathogens and antidiabetic properties. Microb Pathog. 2017, 112, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Aarti, C.; Khusro, A.; Varghese, R.; Arasu, M.V.; Agastian, P.; Al-Dhabi, N.A.; Ilavenil, S.; Choi, K.C. In vitro studies on probiotic and antioxidant properties of Lactobacillus brevis strain LAP2 isolated from Hentak, a fermented fish product of North-East India. LWT Food Sci. Technol. 2017, 86, 438–446. [Google Scholar] [CrossRef]
- Rushdy, A.A.; Gomaa, E.Z. Antimicrobial compounds produced by probiotic Lactobacillus brevis isolated from dairy products. Ann. Microbiol. 2013, 63, 81–90. [Google Scholar] [CrossRef]
- Liu, H.; Gong, J.; Chabot, D.; Miller, S.S.; Cui, S.W.; Ma, J.; Zhong, F.; Wang, Q. Incorporation of polysaccharides into sodium caseinate-low melting point fat microparticles improves probiotic bacterial survival during simulated gastrointestinal digestion and storage. Food Hydrocolloids 2016, 54, 328–337. [Google Scholar] [CrossRef]
- Rönkä, E.; Malinen, E.; Saarela, M.; Rinta-Koski, M.; Aarnikunnas, J.; Palva, A. Probiotic and milk technological properties of Lactobacillus brevis. Int. J. Food Microbiol. 2003, 83, 63–74. [Google Scholar] [CrossRef]
- Uroić, K.; Novak, J.; Hynönen, U.; Pietilä, T.E.; Pavunc, A.L.; Kant, R.; Kos, B.; Palva, A.; Šušković, J. The role of S-layer in adhesive and immunomodulating properties of probiotic starter culture Lactobacillus brevis D6 isolated from artisanal smoked fresh cheese. LWT Food Sci. Technol. 2016, 69, 623–632. [Google Scholar] [CrossRef]
- Delcaru, C.; Alexandru, I.; Podgoreanu, P.; Cristea, V.C.; Bleotu, C.; Chifiriuc, M.C.; Bezirtzoglou, E.; Lazar, V. Antagonistic activities of some Bifidobacterium sp. strains isolated from resident infant gastrointestinal microbiota on Gram-negative enteric pathogens. Anaerobe 2016, 39, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Gilliland, S.; Walker, D. Factors to consider when selecting a culture of Lactobacillus acidophilus as a dietary adjunct to produce a hypocholesterolemic effect in humans. J. Dairy Sci. 1990, 73, 905–911. [Google Scholar] [CrossRef]
- Kimoto-Nira, H.; Suzuki, S.; Suganuma, H.; Moriya, N.; Suzuki, C. Growth characteristics of Lactobacillus brevis KB290 in the presence of bile. Anaerobe 2015, 35, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Barrett, E.; Ross, R.; O’toole, P.; Fitzgerald, G.; Stanton, C. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 2012, 113, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Hartemink, R.; Domenech, V.; Rombouts, F. LAMVAB-a new selective medium for the isolation of lactobacilli from faeces. J. Microbiol. Meth. 1997, 29, 77–84. [Google Scholar] [CrossRef]
- Vanos, V.; Cox, L. Rapid routine method for the detection of lactic acid bacteria among competitive flora. Food Microbiol. 1986, 3, 223–234. [Google Scholar] [CrossRef]
- Zuo, G.C.; Yang, J.Y.; Hao, Y.; Dong, Y.X.; Wu, C.F. Ethanol and acetaldehyde induce similar changes in extracellular levels of glutamate, taurine and GABA in rat anterior cingulate cortex. Toxicol. Lett. 2007, 169, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Cebeci, A.; Gürakan, C. Properties of potential probiotic Lactobacillus plantarum strains. Food Microbiol. 2003, 20, 511–518. [Google Scholar] [CrossRef]

| Medium | MRS | MRS-M | ||||
|---|---|---|---|---|---|---|
| Group | pH Value | Viability (log CFU/mL) | GABA (mg/L) | pH Value | Viability (log CFU/mL) | GABA (mg/L) |
| Amount of MSG (mM) | ||||||
| 0 | N/A | N/A | N/A | 4.27 ± 0.01 | 8.63 ± 0.01 | 773.00 ± 62.23 f |
| 200 | N/A | N/A | N/A | 5.56 ± 0.01 | 8.14 ± 0.02 | 8209.88 ± 326.53 e |
| 400 | N/A | N/A | N/A | 5.57 ± 0.18 | 7.98 ± 0.01 | 11,764.66 ± 4.24 d |
| 450 | N/A | N/A | N/A | 6.20 ± 0.01 | 7.96 ± 0.05 | 13,103.67 ± 477.80 d |
| 500 | N/A | N/A | N/A | 6.72 ± 0.01 | 8.21 ± 0.17 | 13,769.12 ± 852.35 c |
| 550 | N/A | N/A | N/A | 6.71 ± 0.14 | 8.14 ± 0.11 | 15,143.22 ± 182.87 a |
| 600 | N/A | N/A | N/A | 6.70 ± 0.04 | 8.28 ± 0.07 | 14,133.06 ± 113.63 b |
| 650 | N/A | N/A | N/A | 6.75 ± 0.01 | 7.96 ± 0.02 | 14,673.63 ± 455.74 ab |
| Cell inoculum (log CFU/mL) | ||||||
| 7 | 4.41 ± 0.01 a | 8.00 ± 0.01 b | 711.00 ± 0.00 b | 6.85 ± 0.05 b | 9.05 ± 0.06 b | 9058.00 ± 321.00 b |
| 8 | 4.41 ± 0.01 a | 8.21 ± 0.01 a | 739.00 ± 22.63 ab | 6.89 ± 0.01 b | 9.08 ± 0.01 b | 9209.00 ± 313.00 b |
| 9 | 4.43 ± 0.01 a | 8.28 ± 0.04 a | 773.00 ± 9.90 a | 6.99 ± 0.01 a | 9.30 ± 0.03 a | 14,443.00 ± 138.00 a |
| Temperature (°C) | ||||||
| 25 | 4.25 ± 0.21 b | 8.40 ± 0.03 ab | 922.80 ± 103.81 b | 6.94 ± 0.03 ab | 9.06 ± 0.00 a | 17,669.33 ± 1442.80 b |
| 30 | 4.64 ± 0.02 a | 8.66 ± 0.02 a | 1203.62 ± 34.92 a | 6.99 ± 0.10 ab | 9.06 ± 0.03 a | 21,936.27 ± 635.93 a |
| 35 | 4.31 ± 0.00 b | 8.22 ± 0.13 ab | 780.61 ± 108.27 bc | 6.95 ± 0.07 ab | 9.02 ± 0.04 a | 16,839.74 ± 306.42 bc |
| 37 | 4.38 ± 0.03 b | 8.25 ± 0.27 ab | 702.89 ± 9.24 c | 7.05 ± 0.07 a | 9.09 ± 0.01 a | 14,721.66 ± 1028.51 c |
| 40 | 4.34 ± 0.06 b | 8.16 ± 0.23 ab | 341.97 ± 7.20 d | 6.86 ± 0.06 b | 8.72 ± 0.01 b | 10,779.06 ± 1111.58 d |
| 45 | 4.47 ± 0.05 ab | 8.37 ± 0.01 ab | 112.51 ± 8.61 e | 5.68 ± 0.01 c | 7.56 ± 0.02 c | 310.86 ± 8.22 e |
| Initial pHs | ||||||
| 3.5 | 3.33 ± 0.03 c | 5.57 ± 0.04 c | 140.63 ± 15.88 e | 3.19 ± 0.00 f | 7.39 ± 0.11 c | 226.56 ± 11.82 d |
| 4.0 | 4.41 ± 0.11 b | 8.03 ± 0.05 b | 956.71 ± 44.00 ab | 6.52 ± 0.01 b | 8.93 ± 0.03 ab | 24,684.34 ± 562.31 a |
| 4.5 | 4.58 ± 0.04 ab | 8.29 ± 0.01 a | 982.97 ± 44.84 ab | 6.70 ± 0.05 a | 8.90 ± 0.05 b | 25,359.36 ± 541.02 a |
| 5.0 | 4.53 ± 0.04 ab | 8.28 ± 0.09 a | 895.54 ± 17.09 bc | 6.22 ± 0.02 c | 8.88 ± 0.05 b | 21,155.93 ± 336.47 b |
| 5.5 | 4.63 ± 0.10 a | 8.15 ± 0.06 ab | 807.85 ± 46.91 d | 6.18 ± 0.01 cd | 8.98 ± 0.02 ab | 19,883.72 ± 109.77 b |
| 6.0 | 4.64 ± 0.06 a | 8.13 ± 0.19 ab | 818.32 ± 33.38 cd | 6.11 ± 0.03 de | 9.03 ± 0.08 ab | 19,411.73 ± 687.76 b |
| 6.5 | 4.61 ± 0.06 a | 8.25 ± 0.10 ab | 765.96 ± 29.60 d | 6.11 ± 0.07 e | 9.08 ± 0.07 a | 11,473.79 ± 709.65 c |
| pH | Population Changes Compared to Initial Count/Acid Tolerance Time (log CFU/mL) | |||||||
| 0 h | 0.5 h | 1 h | 2 h | 3 h | Survival Rate (%) | |||
| 2 | 7.35 ± 0.01 bA | 7.06 ± 0.05 aAB | 6.47 ± 0.66 aBC | 5.78 ± 0.19 bC | 4.78 ± 0.01 dD | 65 | ||
| 2.5 | 7.34 ± 0.02 bA | 7.10 ± 0.07 aB | 7.04 ± 0.01 aB | 5.96 ± 0.10 bC | 5.69 ± 0.04 cD | 78 | ||
| 3 | 7.79 ± 0.00 aA | 7.11 ± 0.03 aB | 6.97 ± 0.03 aC | 6.82 ± 0.00 aD | 6.12 ± 0.06 bE | 79 | ||
| 7 | 7.74 ± 0.05 aA | 7.11 ± 0.02 aB | 7.11 ± 0.06 aB | 7.11 ± 0.06 aB | 6.92 ± 0.01 aC | 89 | ||
| Bile Salt (%) | Population Changes Compared to Initial Count/Bile Salt Tolerance Time (log CFU/mL) | |||||||
| 0 h | 2 h | 4 h | 6 h | 12 h | 24 h | Survival Rate (%) | ||
| 0 | 7.82 ± 0.05 aA | 7.17 ± 0.13 aB | 6.94 ± 0.03 aBC | 7.04 ± 0.13 aBC | 7.02 ± 0.03 aBC | 7.02 ± 0.03 aC | 90 | |
| 0.15 | 7.82 ± 0.05 aA | 7.08 ± 0.05 abB | 7.14 ± 0.05 aB | 7.09 ± 0.09 aB | 7.08 ± 0.05 aB | 7.10 ± 0.28 aB | 91 | |
| 0.3 | 7.82 ± 0.05 aA | 6.89 ± 0.05 bB | 7.21 ± 0.28 aB | 7.10 ± 0.04 aB | 7.04 ± 0.02 aB | 7.04 ± 0.18 aB | 90 | |
| 0.45 | 7.82 ± 0.05 aA | 7.08 ± 0.09 abB | 7.04 ± 0.07 aCD | 7.05 ± 0.10 aCD | 6.85 ± 0.13 aCD | 6.89 ± 0.04 aD | 88 | |
| Cell Survival Ability | Gastric Juice at Treated Time | Intestinal Juice at Treated Time | ||||||
| 0 h | 1 h | 2 h | 3 h | 2 h | 4 h | 6 h | 12 h | |
| Survival count (log CFU/mL) | 7.34 ± 0.02 A | 6.15 ± 0.01 B | 5.71 ± 0.25 C | 4.89 ± 0.00 D | 4.70 ± 0.06 E | 4.27 ± 0.03 F | 4.17 ± 0.11 F | 4.12 ± 0.18 G |
| Survival rate (%) | 100.0 | 83.8 | 77.8 | 66.6 | 64.00 | 58.00 | 56.80 | 56.10 |
| Strain | Am | Cm | Em | Kan | Sm | Sp | Tc | Vm |
|---|---|---|---|---|---|---|---|---|
| L. brevis RK03 | S(2.0) | S(32.0) | S(16.0) | S(128.0) | R | R | S(32.0) | R |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, C.-H.; Hsueh, Y.-H.; Kuo, J.-M.; Liu, S.-J. Characterization of a Potential Probiotic Lactobacillus brevis RK03 and Efficient Production of γ-Aminobutyric Acid in Batch Fermentation. Int. J. Mol. Sci. 2018, 19, 143. https://doi.org/10.3390/ijms19010143
Wu C-H, Hsueh Y-H, Kuo J-M, Liu S-J. Characterization of a Potential Probiotic Lactobacillus brevis RK03 and Efficient Production of γ-Aminobutyric Acid in Batch Fermentation. International Journal of Molecular Sciences. 2018; 19(1):143. https://doi.org/10.3390/ijms19010143
Chicago/Turabian StyleWu, Chien-Hui, Yi-Huang Hsueh, Jen-Min Kuo, and Si-Jia Liu. 2018. "Characterization of a Potential Probiotic Lactobacillus brevis RK03 and Efficient Production of γ-Aminobutyric Acid in Batch Fermentation" International Journal of Molecular Sciences 19, no. 1: 143. https://doi.org/10.3390/ijms19010143





