Protein Activity of the Fusarium fujikuroi Rhodopsins CarO and OpsA and Their Relation to Fungus–Plant Interaction
Abstract
:1. Introduction
2. Results
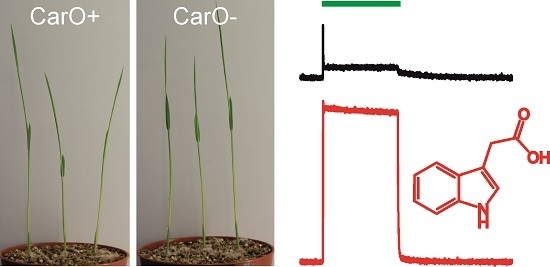
2.1. CarO Pumping Activity Is Enhanced by Indole-3-Acetic Acid (IAA) and Acetate Whereas OpsA Does Not Exhibit Pumping Activity
2.2. Fungal Rhodopsins Are Predominant in Phyto-associated Fungi
2.3. Bakanae Symptoms of Rice Plants Are Affected by the Null CarO Mutation in the Infective Fungus
2.4. Light Exerts Minor Influence on mRNA Levels for Genes of G Proteins
3. Discussion
4. Materials and Methods
4.1. Fungal Strains and Culture Conditions
4.2. Cell Cultures
4.3. Molecular Biology
4.4. Patch-Clamp
4.5. Fluorescence Microscopy
4.6. Rice Plant Infection Assays
4.7. Determination of Chlorophyll and Carotenoid Plant Content
4.8. Gibberellin Production
4.9. Gene Expression Analyses
4.10. Sequence Analyses
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BLAST | Basic Local Alignment Search Tool |
| BR | Bacteriorhodopsin |
| DPSS | Diode Pumped Solid State |
| GA | Gibberellic Acid |
| IAA | Indole-3-Acetic Acid |
| LR | Leptosphaeria Rhodopsin |
| NaAc | Sodium Acetate |
| NaGlu | Sodium Gluconate |
| NR | Neurospora Rhodopsin |
| ORP | Opsin Related Protein |
| SEM | Standard Error of the Mean |
| TM | Transmembrane Helix |
| WOA | Weak Organic Acid |
References
- Turra, D.; Segorbe, D.; Di Pietro, A. Protein kinases in plant-pathogenic fungi: Conserved regulators of infection. Annu. Rev. Phytopathol. 2014, 52, 267–288. [Google Scholar] [CrossRef] [PubMed]
- Fuller, K.K.; Loros, J.J.; Dunlap, J.C. Fungal photobiology: Visible light as a signal for stress, space and time. Curr. Genet. 2015, 61, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Corrochano, L.M.; Galland, P. Photomorphogenesis and Gravitropism in Fungi. In Growth, Differentiation and Sexuality—The Mycota I; Wendland, J., Ed.; Springer: Berlin, Germany, 2016; pp. 235–263. [Google Scholar]
- Corrochano, L.M. Fungal photoreceptors: Sensory molecules for fungal development and behaviour. Photochem. Photobiol. Sci. 2007, 6, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.S. Proton-pumping microbial rhodopsins—Ubiquitous structurally simple helpers of respiration and photosynthesis. Struct. Basis Biol. Energy Gener. 2014, 39, 1–20. [Google Scholar] [CrossRef]
- Brown, L.S.; Dioumaev, A.K.; Lanyi, J.K.; Spudich, E.N.; Spudich, J.L. Photochemical reaction cycle and proton transfers in Neurospora rhodopsin. J. Biol. Chem. 2001, 276, 32495–32505. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Solomon, P.; Oliver, R.P.; Brown, L.S. Photochemical characterization of a novel fungal rhodopsin from Phaeosphaeria nodorum. Biochim. Biophys. Acta Bioenerg. 2011, 1807, 1457–1466. [Google Scholar] [CrossRef] [PubMed]
- García-Martínez, J.; Brunk, M.; Avalos, J.; Terpitz, U. The CarO rhodopsin of the fungus Fusarium fujikuroi is a light-driven proton pump that retards spore germination. Sci. Rep. 2015, 5, 7798. [Google Scholar] [CrossRef] [PubMed]
- Waschuk, S.A.; Bezerra, A.G.; Shi, L.; Brown, L.S. Leptosphaeria rhodopsin: Bacteriorhodopsin-like proton pump from a eukaryote. Proc. Natl. Acad. Sci. USA 2005, 102, 6879–6883. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, J.; Li, N.; Li, J.; Trail, F.; Dunlap, J.C.; Townsend, J.P. Light sensing by opsins and fungal ecology: NOP-1 modulates entry into sexual reproduction in response to environmental cues. Mol. Ecol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Piper, P.W.; Ortiz-Calderon, C.; Holyoak, C.; Coote, P.; Cole, M. Hsp30, the integral plasma membrane heat shock protein of Saccharomyces cerevisiae, is a stress-inducible regulator of plasma membrane H+-ATPase. Cell Stress Chaperones 1997, 2, 12–24. [Google Scholar] [CrossRef]
- Graul, R.C.; Sadée, W. Evolutionary relationships among proteins probed by an iterative neighborhood cluster analysis (INCA). Alignment of bacteriorhodopsins with the yeast sequence YRO2. Pharm. Res. 1997, 14, 1533–1541. [Google Scholar] [CrossRef] [PubMed]
- Bieszke, J.A.; Braun, E.L.; Bean, L.E.; Kang, S.C.; Natvig, D.O.; Borkovich, K.A. The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc. Natl. Acad. Sci. USA 1999, 96, 8034–8039. [Google Scholar] [CrossRef] [PubMed]
- Prado, M.M.; Prado-Cabrero, A.; Fernández-Martín, R.; Avalos, J. A gene of the opsin family in the carotenoid gene cluster of Fusarium fujikuroi. Curr. Genet. 2004, 46, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.S.; Jung, K.H. Bacteriorhodopsin-like proteins of eubacteria and fungi: The extent of conservation of the haloarchaeal proton-pumping mechanism. Photochem. Photobiol. Sci. 2006, 5, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Chow, B.Y.; Han, X.; Dobry, A.S.; Qian, X.F.; Chuong, A.S.; Li, M.J.; Henninger, M.A.; Belfort, G.M.; Lin, Y.X.; Monahan, P.E.; et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature 2010, 463, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Sunder, S. Foot rot and bakanae of rice: An overview. Rev. Plant Pathol. 2012, 5, 565–604. [Google Scholar]
- Avalos, J.; Cerdá-Olmedo, E.; Reyes, F.; Barrero, A.F. Gibberellins and other metabolites of Fusarium fujikuroi and related fungi. Curr. Org. Chem. 2007, 11, 721–737. [Google Scholar] [CrossRef]
- Hedden, P.; Sponsel, V. A Century of gibberellin research. J. Plant Growth Regul. 2015, 34, 740–760. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Solanki, I.S.; Bashyal, B.M.; Singh, Y.; Srivastava, K. Bakanae of rice—An emerging disease in Asia. J. Anim. Plant Sci. 2015, 25, 1499–1514. [Google Scholar]
- Ou, S.H. Bakanae Disease and Food Rot. In Rice Diseases; Common Wealth Mycological Institute: Farnham Royal, UK, 1985; pp. 262–272. [Google Scholar]
- Niehaus, E.-M.; Díaz-Sánchez, V.; von Bargen, K.W.; Kleigrewe, K.; Humpf, H.-U.; Limón, M.C.; Tudzynski, B. Fusarins and Fusaric Acid in Fusaria; Springer: New York, NY, USA, 2014; pp. 239–262. [Google Scholar]
- Estrada, A.F.; Avalos, J. Regulation and targeted mutation of opsA, coding for the NOP-1 opsin orthologue in Fusarium fujikuroi. J. Mol. Biol. 2009, 387, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Brunk, M.; Sputh, S.; Doose, S.; van der Linde, S.; Terpitz, U. HyphaTracker: An ImageJ toolbox for time-resolved analysis of spore germination in filamentous fungi. Sci. Rep. 2018. [Google Scholar] [CrossRef]
- Atamna-Ismaeel, N.; Finkel, O.M.; Glaser, F.; Sharon, I.; Schneider, R.; Post, A.F.; Spudich, J.L.; von Mering, C.; Vorholt, J.A.; Iluz, D.; et al. Microbial rhodopsins on leaf surfaces of terrestrial plants. Environ. Microbiol. 2012, 14, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Ohm, R.A.; Grigoriev, I.V.; Srivastava, A. Fungal-specific transcription factor AbPf2 activates pathogenicity in Alternaria brassicicola. Plant J. 2013, 75, 498–514. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A. Small heat shock proteins (HSP12, HSP20 and HSP30) play a role in Ustilago maydis pathogenesis. FEMS Microbiol. Lett. 2014, 361, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Caracuel, Z.; Roncero, M.I.G.; Espeso, E.A.; González-Verdejo, C.I.; García-Maceira, F.I.; Di Pietro, A. The pH signalling transcription factor PacC controls virulence in the plant pathogen Fusarium oxysporum. Mol. Microbiol. 2003, 48, 765–779. [Google Scholar] [CrossRef] [PubMed]
- Alkan, N.; Espeso, E.A.; Prusky, D. Virulence regulation of phytopathogenic fungi by pH. Antioxid. Redox Signal. 2013, 19, 1012–1025. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, R.; Kesselmeier, J.; Planck, M.; Box, P.O. Apoplastic solute concentrations of organic acids and mineral nutrients in the leaves of several fagaceae. Plant Cell Physiol. 1999, 40, 604–612. [Google Scholar] [CrossRef]
- Capurro, V.; Gianotti, A.; Caci, E.; Ravazzolo, R.; Galietta, L.J.V.; Zegarra-Moran, O. Functional analysis of acid-activated Cl− channels: Properties and mechanisms of regulation. Biochim. Biophys. Acta Biomembr. 2015, 1848, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Bieszke, J.A.; Spudich, E.N.; Scott, K.L.; Borkovich, K.A.; Spudich, J.L. A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry 1999, 38, 14138–14145. [Google Scholar] [CrossRef] [PubMed]
- Sakmar, T.P.; Franke, R.R.; Khorana, H.G. The role of the retinylidene Schiff base counterion in rhodopsin in determining wavelength absorbance and Schiff base pKa. Proc. Natl. Acad. Sci. USA 1991, 88, 3079–3083. [Google Scholar] [CrossRef] [PubMed]
- Váró, G.; Lanyi, J.K. Photoreactions of bacteriorhodopsin at acid pH. Biophys. J. 1989, 56, 1143–1151. [Google Scholar] [CrossRef]
- Stergiopoulos, I.; Kourmpetis, Y.A.I.; Slot, J.C.; Bakker, F.T.; De Wit, P.J.G.M.; Rokas, A. In silico characterization and molecular evolutionary analysis of a novel superfamily of fungal effector proteins. Mol. Biol. Evol. 2012, 29, 3371–3384. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Sung, G.-H.; Lopez-Giraldez, F.; Townsend, J.P.; Miadlikowska, J.; Hofstetter, V.; Robbertse, B.; Matheny, P.B.; Kauff, F.; Wang, Z.; et al. The ascomycota tree of life: A phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst. Biol. 2009, 58, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-Y.; Seo, J.-A.; Kim, J.-E.; Han, K.-H.; Shim, W.-B.; Yun, S.-H.; Lee, Y.-W. Functional analyses of heterotrimeric G protein Gα and Gβ subunits in Gibberella zeae. Microbiology 2008, 154, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Avalos, J.; Nordzieke, S.; Parra, O.; Pardo-Medina, J.; Limón, M.C. Carotenoid Production by Filamentous Fungi and Yeasts. In Biotechnology of Yeasts and Filamentous Fungi; Sibirny, A.A., Ed.; Springer International Publishing: Cham, Germany, 2017; pp. 225–279. ISBN 978-3-319-58829-2. [Google Scholar]
- Niehaus, E.-M.; Kim, H.-K.; Münsterkötter, M.; Janevska, S.; Arndt, B.; Kalinina, S.A.; Houterman, P.M.; Ahn, I.-P.; Alberti, I.; Tonti, S.; et al. Comparative genomics of geographically distant Fusarium fujikuroi isolates revealed two distinct pathotypes correlating with secondary metabolite profiles. PLoS Pathog. 2017, 13, e1006670. [Google Scholar] [CrossRef] [PubMed]
- Kihara, J.; Tanaka, N.; Ueno, M.; Arase, S. Cloning and expression analysis of two opsin-like genes in the phytopathogenic fungus Bipolaris oryzae. FEMS Microbiol. Lett. 2009, 295, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.; Shen, C.; Fu, Y.; Xie, J.; Jiang, D.; Li, G.; Cheng, J. The microbial opsin homolog Sop1 is involved in Sclerotinia sclerotiorum development and environmental stress response. Front. Microbiol. 2015, 6, 1504. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.C.; Hui, S.; Choi, J.; Asiegbu, F.O.; Valkonen, J.P.; Lee, Y.H. Secret lifestyles of Neurospora crassa. Sci. Rep. 2014, 4, 5135. [Google Scholar] [CrossRef] [PubMed]
- Wulff, E.G.; Sørensen, J.L.; Lübeck, M.; Nielsen, K.F.; Thrane, U.; Torp, J. Fusarium spp. associated with rice Bakanae: Ecology, genetic diversity, pathogenicity and toxigenicity. Environ. Microbiol. 2010, 12, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Wiemann, P.; Sieber, C.M.K.; von Bargen, K.W.; Studt, L.; Niehaus, E.-M.; Espino, J.J.; Huß, K.; Michielse, C.B.; Albermann, S.; Wagner, D.; et al. Deciphering the cryptic genome: Genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013, 9, e1003475. [Google Scholar] [CrossRef] [PubMed]
- Braga, G.U.L.; Rangel, D.E.N.; Fernandes, É.K.K.; Flint, S.D.; Roberts, D.W. Molecular and physiological effects of environmental UV radiation on fungal conidia. Curr. Genet. 2015, 61, 405–425. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.E.M.; Dillon, R.J.; Charnley, A.K. Influence of accelerated germination of conidia on the pathogenicity of Metarhizium anisopliae for Manduca sexta. J. Invertebr. Pathol. 1989, 54, 277–279. [Google Scholar] [CrossRef]
- Aver’yanov, A.A.; Lapikova, V.P.; Pasechnik, T.D.; Abramova, O.S.; Gaivoronskaya, L.M.; Kuznetsov, V.V.; Baker, C.J. Pre-illumination of rice blast conidia induces tolerance to subsequent oxidative stress. Fungal Biol. 2014, 118, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Canessa, P.; Schumacher, J.; Hevia, M.A.; Tudzynski, P.; Larrondo, L.F. Assessing the effects of light on differentiation and virulence of the plant pathogen Botrytis cinerea: Characterization of the white collar complex. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.M.; Ramkumar, G.; Lee, Y.H. Light quality influences the virulence and physiological responses of Colletotrichum acutatum causing anthracnose in pepper plants. J. Appl. Microbiol. 2013, 115, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Loayza, P.; White, J.F., Jr.; Torres, M.S.; Balslev, H.; Kristiansen, T.; Svenning, J.C.; Gil, N. Light converts endosymbiotic fungus to pathogen, influencing seedling survival and niche-space filling of a common tropical tree, Iriartea deltoidea. PLoS ONE 2011, 6, e16386. [Google Scholar] [CrossRef] [PubMed]
- Borkovich, K.A.; Ebbole, D.J. Cellular & Molecular Biology of Filamentous Fungi; Borkovich, K.A., Ed.; American Society for Microbiology: Washington, DC, USA, 2010; ISBN 9786613034342. [Google Scholar]
- Spudich, J.L.; Sineshchekov, O.A.; Govorunova, E.G. Mechanism divergence in microbial rhodopsins. Biochim. Biophys. Acta 2014, 1837, 546–552. [Google Scholar] [CrossRef] [PubMed]
- Lórenz-Fonfría, V.A.; Heberle, J. Channelrhodopsin unchained: Structure and mechanism of a light-gated cation channel. Biochim. Biophys. Acta Bioenerg. 2014, 1837, 626–642. [Google Scholar] [CrossRef] [PubMed]
- Nigovic, B.; KojicProdic, B.; Antolic, S.; Tomic, S.; Puntarec, V.; Cohen, J.D. Structural studies on monohalogenated derivatives of the phytohormone indole-3-acetic acid (auxin). Acta Crystallogr. Sect. B 1996, 52, 332–343. [Google Scholar] [CrossRef]
- Nielsen, J. Physiological Engineering Aspects of Penicillium Chrysogenum; World Scientific: Singapore, 1997; ISBN 978-981-02-2765-4. [Google Scholar]
- Bean, R.C.; Shepherd, W.C.; Chan, H. Permeability of lipid bilayer membranes to organic solutes. J. Gen. Physiol. 1968, 52, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Stewart, A.K.; Chernova, M.N.; Kunes, Y.Z.; Alper, S.L. Regulation of AE2 anion exchanger by intracellular pH: Critical regions of the NH2-terminal cytoplasmic domain. Am. J. Physiol. 2001, 281, C1344–C1354. [Google Scholar] [CrossRef] [PubMed]
- Lörinczi, E.; Verhoefen, M.K.; Wachtveitl, J.; Woerner, A.C.; Glaubitz, C.; Engelhard, M.; Bamberg, E.; Friedrich, T. Voltage- and pH-dependent changes in vectoriality of photocurrents mediated by wild-type and mutant proteorhodopsins upon expression in Xenopus oocytes. J. Mol. Biol. 2009, 393, 320–341. [Google Scholar] [CrossRef] [PubMed]
- Terpitz, U.; Raimunda, D.; Westhoff, M.; Sukhorukov, V.L.; Beaugé, L.; Bamberg, E.; Zimmermann, D.; Beauge, L.; Bamberg, E.; Zimmermann, D. Electrofused giant protoplasts of Saccharomyces cerevisiae as a novel system for electrophysiological studies on membrane proteins. Biochim. Biophys. Acta. Biomembr. 2008, 1778, 1493–1500. [Google Scholar] [CrossRef] [PubMed]
- Lanyi, J.K. Halorhodopsin: A light-driven chloride ion pump. Annu. Rev. Biophys. Biophys. Chem. 1986, 15, 11–28. [Google Scholar] [CrossRef] [PubMed]
- Tsavkelova, E.; Oeser, B.; Oren-Young, L.; Israeli, M.; Sasson, Y.; Tudzynski, B.; Sharon, A. Identification and functional characterization of indole-3-acetamide-mediated IAA biosynthesis in plant-associated Fusarium species. Fungal Genet. Biol. 2012, 49, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Quazi, S.A.J.; Meon, S.; Jaafar, H.; Ahmad, Z.A.B.M. The role of phytohormones in relation to bakanae disease development and symptoms expression. Physiol. Mol. Plant Pathol. 2015, 90, 27–38. [Google Scholar] [CrossRef]
- Terakita, A. The opsins. Genome Biol. 2005, 6, 213. [Google Scholar] [CrossRef] [PubMed]
- Palczewski, K. G Protein–coupled receptor rhodopsin. Annu. Rev. Biochem. 2006, 75, 743–767. [Google Scholar] [CrossRef] [PubMed]
- Avalos, J.; Casadesús, J.; Cerdá-Olmedo, E. Gibberella fujikuroi mutants obtained with UV radiation and N-methyl-N’-nitro-N-nitrosoguanidine. Appl. Environ. Microbiol. 1985, 49, 187–191. [Google Scholar] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Kraehmer, H.; Thomas, C.; Vidotto, F. Rice Production in Europe. In Rice Production Worldwide; Chauhan, B.S., Jabran, K., Mahajan, G., Eds.; Springer International Publishing: Cham, Germany, 2017; pp. 93–116. ISBN 978-3-319-47516-5. [Google Scholar]
- Rigaud, J.; Puppo, A. Indole-3-acetic acid catabolism by soybean bacteroids. J. Gen. Microbiol. 1975, 88, 223–228. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Buschmann, C. Chlorophylls and carotenoids: Measurement and characterization by UV-VIS. Curr. Protoc. Food Anal. Chem. 2001. [Google Scholar] [CrossRef]
- Geissman, T.A.; Verbiscar, A.J.; Phinney, B.O.; Cragg, G. Studies on the biosynthesis of gibberellins from (−)-kaurenoic acid in cultures of Gibberella fujikuroi. Phytochemistry 1966, 5, 933–947. [Google Scholar] [CrossRef]
- Candau, R.; Avalos, J.; Cerdá-Olmedo, E. Gibberellins and carotenoids in the wild type and mutants of Gibberella fujikuroi. Appl. Environ. Microbiol. 1991, 57, 3378–3382. [Google Scholar] [PubMed]
- Di Tommaso, P.; Moretti, S.; Xenarios, I.; Orobitg, M.; Montanyola, A.; Chang, J.-M.; Taly, J.-F.; Notredame, C. T-Coffee: A web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011, 39, W13–W17. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- BLAST:_ Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 15 October 2017).
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- NJplot. Available online: http://doua.prabi.fr/software/njplot (accessed on 15 October 2017).






| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| FFUJ_11802 (carRA) | CAGAAGCTGTTCCCGAAGACA | TGCGATGCCCATTTCTTGA |
| FFUJ_11803 (carB) | TCGGTGTCGAGTACCGTCTCT | TGCCTTGCCGGTTGCTT |
| FFUJ_11804 (carO) | TGGGCAACGCAGTGACAT | TGCGCAGACAGCCCAGTA |
| FFUJ_04487 | CAACTACCGGCCAACTGTCT | TCTGCATGTGCCTTGTTCTC |
| FFUJ_05248 | TTCGGAAGCTTGCAACAACG | TCGGTGGGTTGATTCGTGAG |
| FFUJ_06643 | CAGCTATCCTGCAGAAGCGA | CATGCTCATCGCCGAAAAGG |
| FFUJ_07379 | TAACCCCGACAACGAGAAAC | GTCTACCCACAGGGCTTTGA |
| FFUJ_08667 | GATGTCCTCCGATCTCGTGT | CTTTCGCTCGGATCTTTGAC |
| FFUJ_09550 (Gβ type) | ATCACCTCGGTGGCTACATC | ATGTCCCAAACCTTGCACTC |
| FFUJ_03226 (Gγ type) | ACCGAGCTCAACAATCGTCT | TGCAGTAGGCAATGATGCTC |
| FFUJ_04397 (tubulin β) | CCGGTGCTGGAAACAACTG | CGAGGACCTGGTCGACAAGT |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adam, A.; Deimel, S.; Pardo-Medina, J.; García-Martínez, J.; Konte, T.; Limón, M.C.; Avalos, J.; Terpitz, U. Protein Activity of the Fusarium fujikuroi Rhodopsins CarO and OpsA and Their Relation to Fungus–Plant Interaction. Int. J. Mol. Sci. 2018, 19, 215. https://doi.org/10.3390/ijms19010215
Adam A, Deimel S, Pardo-Medina J, García-Martínez J, Konte T, Limón MC, Avalos J, Terpitz U. Protein Activity of the Fusarium fujikuroi Rhodopsins CarO and OpsA and Their Relation to Fungus–Plant Interaction. International Journal of Molecular Sciences. 2018; 19(1):215. https://doi.org/10.3390/ijms19010215
Chicago/Turabian StyleAdam, Alexander, Stephan Deimel, Javier Pardo-Medina, Jorge García-Martínez, Tilen Konte, M. Carmen Limón, Javier Avalos, and Ulrich Terpitz. 2018. "Protein Activity of the Fusarium fujikuroi Rhodopsins CarO and OpsA and Their Relation to Fungus–Plant Interaction" International Journal of Molecular Sciences 19, no. 1: 215. https://doi.org/10.3390/ijms19010215