miR-3189-3p Mimics Enhance the Effects of S100A4 siRNA on the Inhibition of Proliferation and Migration of Gastric Cancer Cells by Targeting CFL2
Abstract
:1. Introduction
2. Results
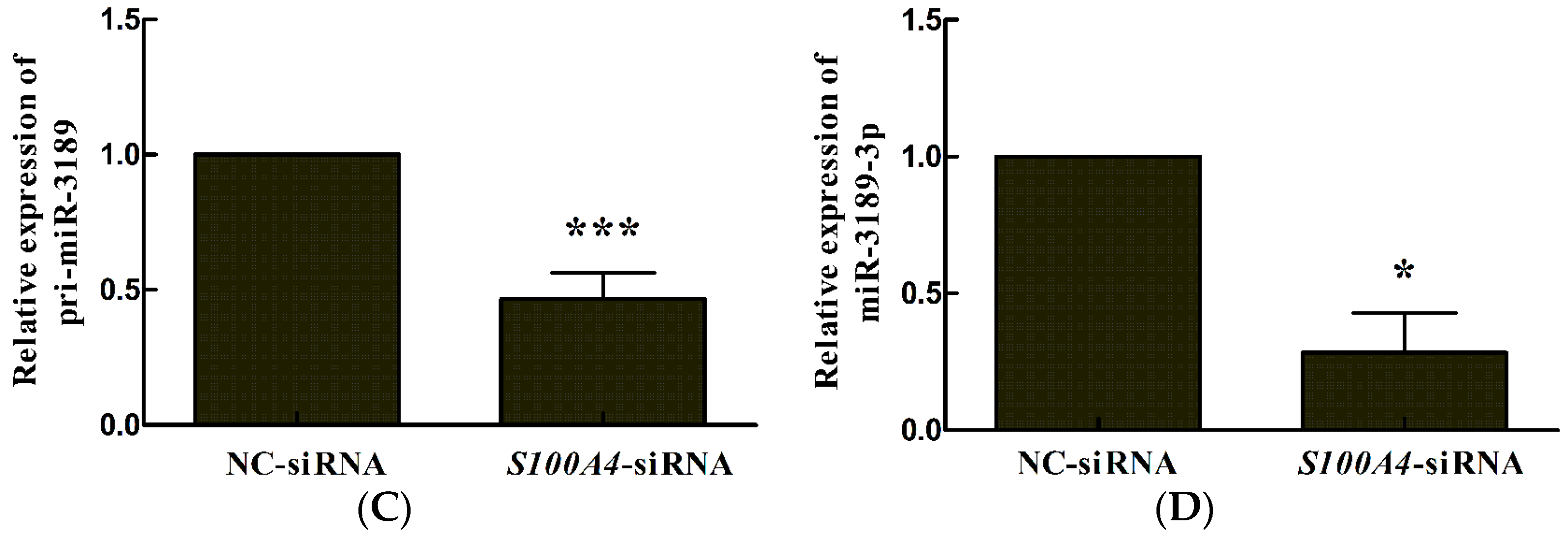
2.1. S100A4 Knockdown Leads to Decreased Expression of miR-3189-3p in MGC803 Cells
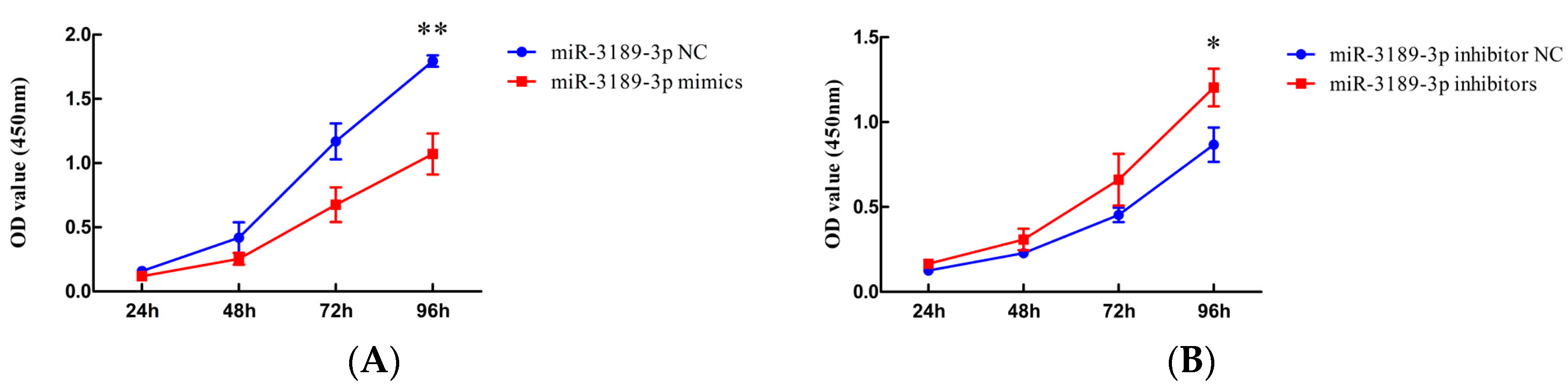
2.2. miR-3189-3p Inhibits the Proliferation of MGC803 Cells
2.3. miR-3189-3p Retards MGC803 Cells Migration
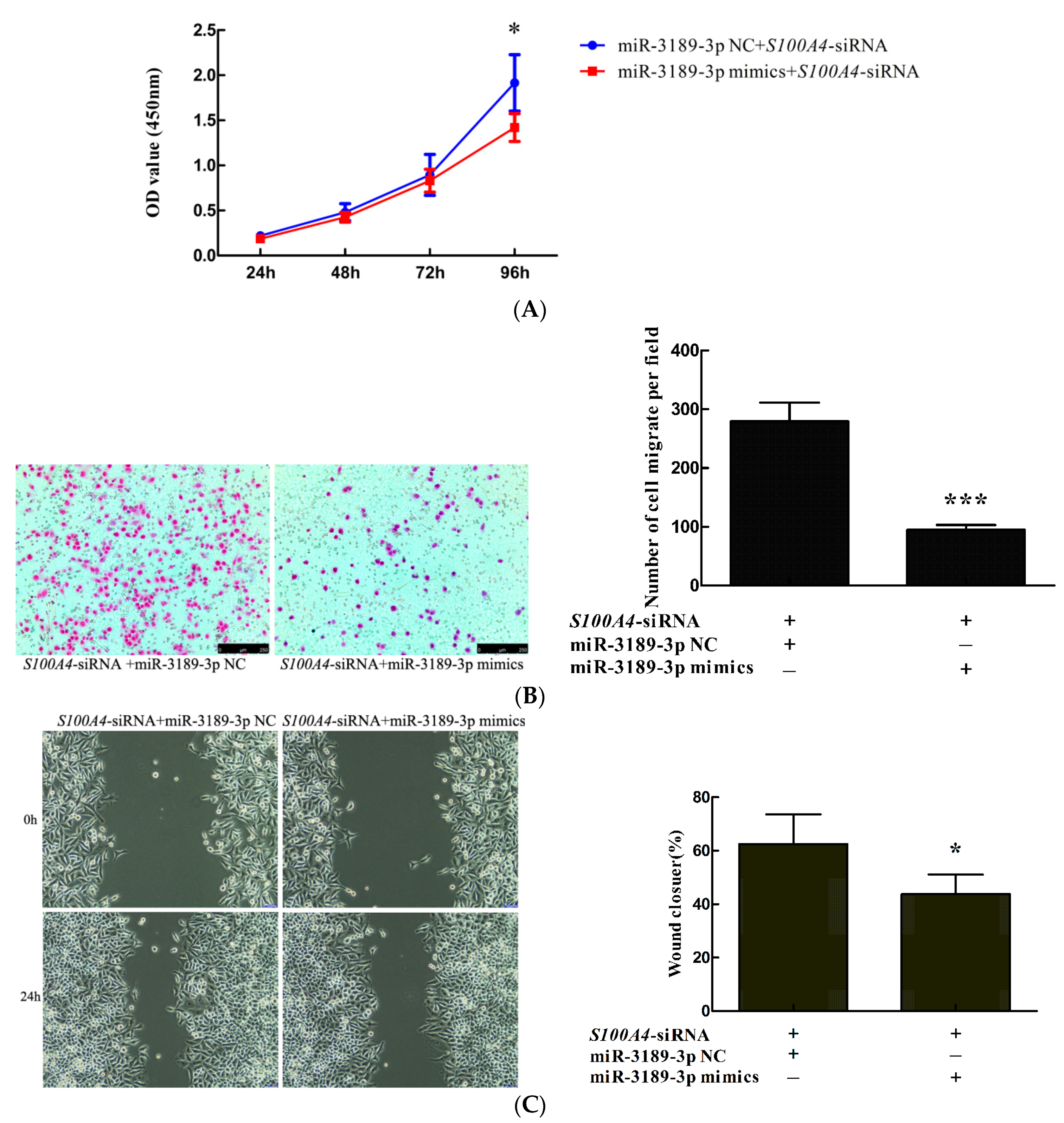
2.4. miR-3189-3p Mimics Enhanced the Effects of S100A4 siRNA on the Inhibition of Proliferation and Migration of MGC803 Cells
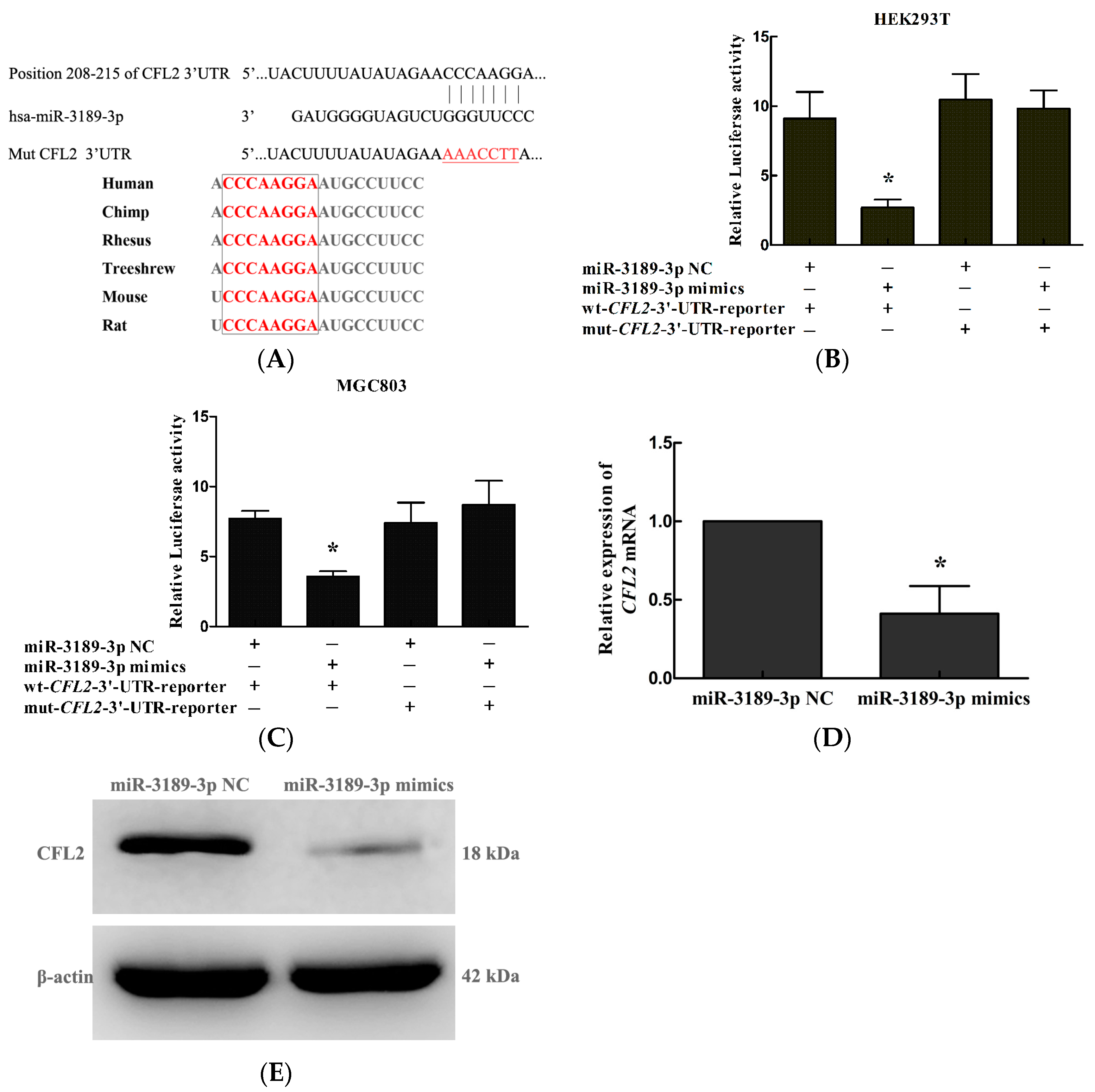
2.5. CFL2 Is a Direct Target Gene of miR-3189-3p in MGC803 Cells
2.6. CFL2 siRNA Inhibits the Proliferation and Migration of MGC803 Cells
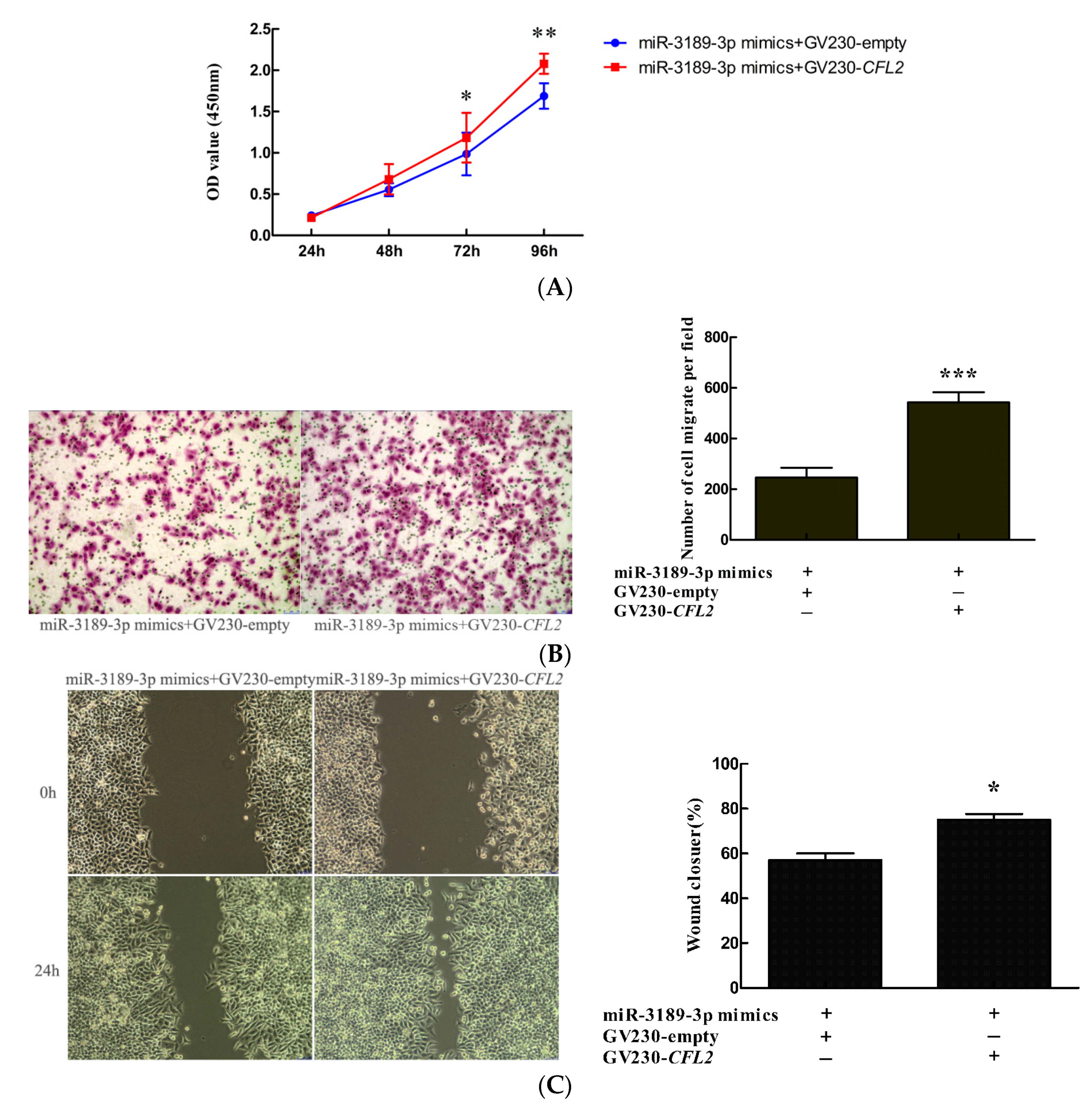
2.7. CFL2 Mediates the Functional Effects of miR-3189-3p on MGC803 Cells.
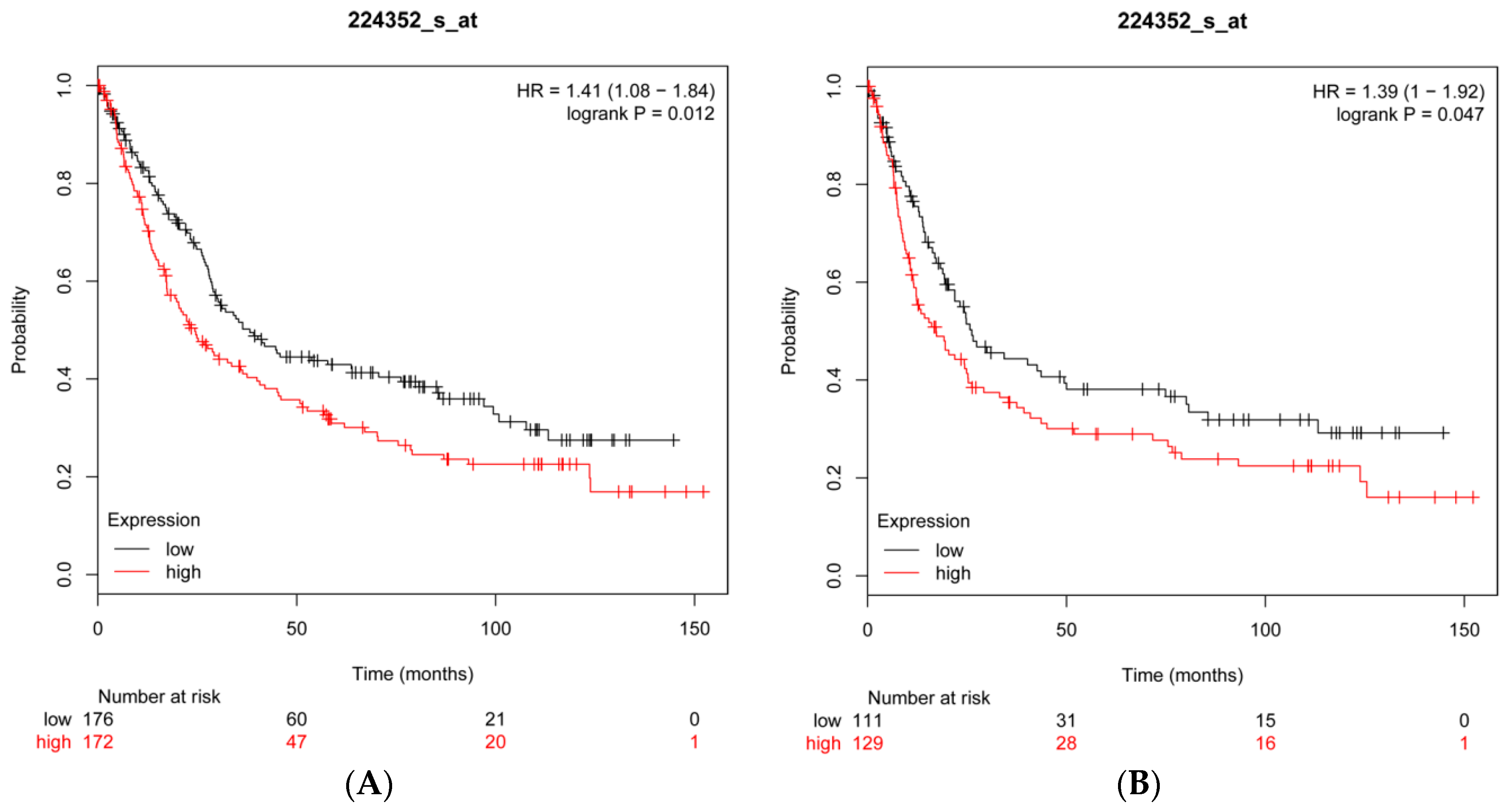
2.8. CFL2 Is an Unfavorable Prognostic Factor for Gastric Cancer
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Transfection
4.3. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
4.4. Western Blotting Analysis
4.5. Dual Luciferase Reporter Assay
4.6. Cell Proliferation
4.7. Transwell Assay
4.8. Wound Healing Assay
4.9. Kaplan–Meier Plotter
4.10. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| GC | gastric cancer |
| miRNA | microRNA |
| 3′-UTR | 3′-untranslated region |
| CFL2 | cofilin 2 |
| OS | overall survival |
| FP | first progression |
| HR | hazard ratio |
| 95%CI | 95% confidence intervals |
| IBMS | Institute of Basic Medical Sciences |
| CAMS | Chinese Academy of Medical Sciences |
| PUMC | Peking Union Medical College |
| SPSS | Statistical Package for the Social Sciences |
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Li, R. Clinicopathological and prognostic value of S100A4 expression in gastric cancer: A meta-analysis. Int. J. Biol. Markers 2014, 29, e99–e111. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Sato, D.; Saiki, Y.; Sunamura, M.; Fukushige, S.; Horii, A. S100A4 is frequently overexpressed in lung cancer cells and promotes cell growth and cell motility. Biochem. Biophys. Res. Commun. 2014, 447, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Wang, L.; Cheng, C.; Du, C.; Lu, X.; Wang, G.; Liu, J. Increased expressions of SATB1 and S100A4 are associated with poor prognosis in human colorectal carcinoma. APMIS 2015, 123, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Liu, J.; Yang, B.; Gao, X.; Gao, L.L.; Kong, Q.Y.; Zhang, P.; Li, H. Inversed Expression Patterns of S100A4 and E-Cadherin in Cervical Cancers: Implication in Epithelial-Mesenchymal Transition. Anat. Rec. 2017, 300, 2184–2191. [Google Scholar] [CrossRef] [PubMed]
- Ismail, T.M.; Bennett, D.; Platt-Higgins, A.M.; Al-Medhity, M.; Barraclough, R.; Rudland, P.S. S100A4 Elevation Empowers Expression of Metastasis Effector Molecules in Human Breast Cancer. Cancer Res. 2017, 77, 780–789. [Google Scholar] [CrossRef] [PubMed]
- Hua, J.; Chen, D.; Fu, H.; Zhang, R.; Shen, W.; Liu, S.; Sun, K.; Sun, X. Short hairpin RNA-mediated inhibition of S100A4 promotes apoptosis and suppresses proliferation of BGC823 gastric cancer cells in vitro and in vivo. Cancer Lett. 2010, 292, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Chen, D.; Fu, H.; Liu, S.; Sun, K.; Sun, X. S100A4 protects gastric cancer cells from anoikis through regulation of alphav and α5 integrin. Cancer Sci. 2011, 102, 1014–1018. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhang, R.; Shen, W.; Fu, H.; Liu, S.; Sun, K.; Sun, X. RPS12-specific shRNA inhibits the proliferation, migration of BGC823 gastric cancer cells with S100A4 as a downstream effector. Int. J. Oncol. 2013, 42, 1763–1769. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Bian, Y.; Wang, Y.; Chen, L.; Yu, A.; Sun, X. S100A4 influences cancer stem cell-like properties of MGC803 gastric cancer cells by regulating GDF15 expression. Int. J. Oncol. 2016, 49, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Huang, S.X.; Zhang, F.; Li, S.J.; Liu, C.; Xi, Y.Y.; Wang, L.; Wang, X.; He, Q.Q.; Sun, C.C.; et al. MicroRNAs: A novel potential biomarker for diagnosis and therapy in patients with non-small cell lung cancer. Cell Prolif. 2017, 50, e12394. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Hayashi, N.; Kuroda, Y.; Ito, S.; Eguchi, H.; Mimori, K. MicroRNAs as Biomarkers in Colorectal Cancer. Cancers 2017, 9, 124. [Google Scholar] [CrossRef] [PubMed]
- Markopoulos, G.S.; Roupakia, E.; Tokamani, M.; Chavdoula, E.; Hatziapostolou, M.; Polytarchou, C.; Marcu, K.B.; Papavassiliou, A.G.; Sandaltzopoulos, R.; Kolettas, E. A step-by-step microRNA guide to cancer development and metastasis. Cell. Oncol. 2017, 40, 303–339. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Tang, W.M.; Zhang, H.; Li, Y.Q.; Peng, Y.; Wang, J.; Liu, G.N.; Huang, X.T.; Zhao, J.J.; Li, G.; et al. miR-646 inhibited cell proliferation and EMT-induced metastasis by targeting FOXK1 in gastric cancer. Br. J. Cancer 2017, 117, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Huang, S.; Long, D. miR-381 inhibits migration and invasion in human gastric carcinoma through downregulatedting SOX4. Oncol. Lett. 2017, 14, 3760–3766. [Google Scholar] [CrossRef] [PubMed]
- Qiao, W.; Cao, N.; Yang, L. MicroRNA-154 inhibits the growth and metastasis of gastric cancer cells by directly targeting MTDH. Oncol. Lett. 2017, 14, 3268–3274. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Jia, B.; Wang, Y.; Wan, S. miR-133b acts as a tumor suppressor and negatively regulates ATP citrate lyase via PPARgamma in gastric cancer. Oncol. Rep. 2017, 38, 3220–3226. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.H.; Li, B.S.; Liu, J.J.; Xiao, Y.F.; Yong, X.; Wang, S.M.; Wu, Y.Y.; Zhu, H.B.; Wang, D.X.; Yang, S.M. miR-93-5p/IFNAR1 axis promotes gastric cancer metastasis through activating the STAT3 signaling pathway. Cancer Lett. 2017, 408, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.F.; Li, X.L.; Subramanian, M.; Shabalina, S.A.; Hara, T.; Zhu, Y.; Huang, J.; Yang, Y.; Wakefield, L.M.; Prasanth, K.V.; et al. Growth differentiation factor-15 encodes a novel microRNA 3189 that functions as a potent regulator of cell death. Cell Death Differ. 2015, 22, 1641–1653. [Google Scholar] [CrossRef] [PubMed]
- Jeansonne, D.; DeLuca, M.; Marrero, L.; Lassak, A.; Pacifici, M.; Wyczechowska, D.; Wilk, A.; Reiss, K.; Peruzzi, F. Anti-tumoral effects of miR-3189-3p in glioblastoma. J. Biol. Chem. 2015, 290, 8067–8080. [Google Scholar] [CrossRef] [PubMed]
- Voigtlander, T.; Gupta, S.K.; Thum, S.; Fendrich, J.; Manns, M.P.; Lankisch, T.O.; Thum, T. MicroRNAs in Serum and Bile of Patients with Primary Sclerosing Cholangitis and/or Cholangiocarcinoma. PLoS ONE 2015, 10, e0139305. [Google Scholar] [CrossRef] [PubMed]
- Sivadas, V.P.; George, N.A.; Kattoor, J.; Kannan, S. Novel mutations and expression alterations in SMAD3/TGFBR2 genes in oral carcinoma correlate with poor prognosis. Genes Chromosomes Cancer 2013, 52, 1042–1052. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, A.A.; Ziada, M.; Elshaikh, A.; Datta, A.; Kim, H.; Moroz, K.; Srivastav, S.; Thomas, R.; Silberstein, J.L.; Moparty, K.; et al. Identification of microRNA signature and potential pathway targets in prostate cancer. Exp. Biol. Med. 2017, 242, 536–546. [Google Scholar] [CrossRef] [PubMed]
- Radfar, M.H.; Wong, W.; Morris, Q. Computational prediction of intronic microRNA targets using host gene expression reveals novel regulatory mechanisms. PLoS ONE 2011, 6, e19312. [Google Scholar] [CrossRef] [PubMed]
- Bohlig, L.; Friedrich, M.; Engeland, K. p53 activates the PANK1/miRNA-107 gene leading to downregulation of CDK6 and p130 cell cycle proteins. Nucleic Acids Res. 2011, 39, 440–453. [Google Scholar] [CrossRef] [PubMed]
- Baskerville, S.; Bartel, D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005, 11, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Bian, Y.; Wang, Y.; Chen, L.; Yu, A.; Sun, X. FAM107B is regulated by S100A4 and mediates the effect of S100A4 on the proliferation and migration of MGC803 gastric cancer cells. Cell Biol. Int. 2017, 41, 1103–1109. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Xu, Y.; Cai, G.; Guan, Z.; Cai, S. Downregulation of S100A4 expression by RNA interference suppresses cell growth and invasion in human colorectal cancer cells. Oncol. Rep. 2012, 27, 917–922. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Yu, M.; Hao, F.; Dong, A.; Chen, D. Knockdown of S100A4 blocks growth and metastasis of anaplastic thyroid cancer cells in vitro and in vivo. Cancer Biomark. 2016, 17, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chen, D.; Shen, W.; Chen, L.; Yu, A.; Fu, H.; Sun, K.; Sun, X. EZH2 Mediates the Regulation of S100A4 on E-cadherin Expression and the Proliferation, Migration of Gastric Cancer Cells. Hepato-Gastroenterology 2015, 62, 737–741. [Google Scholar] [PubMed]
- Zhang, H.Y.; Zheng, X.Z.; Wang, X.H.; Xuan, X.Y.; Wang, F.; Li, S.S. S100A4 mediated cell invasion and metastasis of esophageal squamous cell carcinoma via the regulation of MMP-2 and E-cadherin activity. Mol. Biol. Rep. 2012, 39, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.L.; Jia, Y.L.; Chen, L.; Zeng, Q.; Zhou, J.N.; Fu, C.J.; Chen, H.X.; Yuan, H.F.; Li, Z.W.; Shi, L.; et al. Hepatocellular carcinoma-associated mesenchymal stem cells promote hepatocarcinoma progression: Role of the S100A4-miR155-SOCS1-MMP9 axis. Hepatology 2013, 57, 2274–2286. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ge, M.; Yin, Y.; Luo, M.; Chen, D. Silencing expression of ribosomal protein L26 and L29 by RNA interfering inhibits proliferation of human pancreatic cancer PANC-1 cells. Mol. Cell. Biochem. 2012, 370, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Yehl, J.; Kudryashova, E.; Reisler, E.; Kudryashov, D.; Polenova, T. Structural Analysis of Human Cofilin 2/Filamentous Actin Assemblies: Atomic-Resolution Insights from Magic Angle Spinning NMR Spectroscopy. Sci. Rep. 2017, 7, 44506. [Google Scholar] [CrossRef] [PubMed]
- Schwickert, A.; Weghake, E.; Bruggemann, K.; Engbers, A.; Brinkmann, B.F.; Kemper, B.; Seggewiss, J.; Stock, C.; Ebnet, K.; Kiesel, L.; et al. microRNA miR-142-3p Inhibits Breast Cancer Cell Invasiveness by Synchronous Targeting of WASL, Integrin Alpha V, and Additional Cytoskeletal Elements. PLoS ONE 2015, 10, e0143993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, D.; Wilson, J.M.; Harvel, N.; Liu, J.; Pei, L.; Huang, S.; Hawthorn, L.; Shi, H. A systematic evaluation of miRNA:mRNA interactions involved in the migration and invasion of breast cancer cells. J. Transl. Med. 2013, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kuramitsu, Y.; Ueno, T.; Suzuki, N.; Yoshino, S.; Iizuka, N.; Zhang, X.; Oka, M.; Nakamura, K. Differential expression of up-regulated cofilin-1 and down-regulated cofilin-2 characteristic of pancreatic cancer tissues. Oncol. Rep. 2011, 26, 1595–1599. [Google Scholar] [PubMed]
- Erkutlu, I.; Cigiloglu, A.; Kalender, M.E.; Alptekin, M.; Demiryurek, A.T.; Suner, A.; Ozkaya, E.; Ulasli, M.; Camci, C. Correlation between Rho-kinase pathway gene expressions and development and progression of glioblastoma multiforme. Tumour Biol. 2013, 34, 1139–1144. [Google Scholar] [CrossRef] [PubMed]
- Szasz, A.M.; Lanczky, A.; Nagy, A.; Forster, S.; Hark, K.; Green, J.E.; Boussioutas, A.; Busuttil, R.; Szabo, A.; Gyorffy, B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1065 patients. Oncotarget 2016, 7, 49322–49333. [Google Scholar] [CrossRef] [PubMed]



| Name of Short Nucleotides | Sequences |
|---|---|
| S100A4-siRNA | 5′-GCAUCGCCAUGAUGUGUAATT-3′ |
| 5′-UUACACAUCAUGGCGAUGCTT-3′ | |
| CFL2-siRNA | 5′-GCAAGUAAAUGGCUUGGAUTT-3′ |
| 5′-AUCCAAGCCAUUUACUUGCTT-3′ | |
| hsa-miR-3189-3p mimics | 5′-CCCUUGGGUCUGAUGGGGUAG-3′ |
| 5′-ACCCCAUCAGACCCAAGGGUU-3′ | |
| Negative Control (NC) | 5′-UUCUCCGAACGUGUCACGUTT-3′ |
| 5′-ACGUGACACGUUCGGAGAATT-3′ | |
| hsa-miR-3189-3p inhibitor | 5′-CUACCCCAUCAGACCCAAGGG-3′ |
| hsa-miR-3189-3p inhibitor NC | 5’-CAGUACUUUUGUGUAGUACAA-3’ |
| Gene | Primer Sequence (5′–3′) |
|---|---|
| S100A4 | F: CCCTGGATGTGATGGTGT |
| R: GTTGTCCCTGTTGCTGTC | |
| GDF15 | F: CTCCAGATTCCGAGAGTTGC |
| R: AGAGATACGCAGGTGCAGGT | |
| pri-miR-3189 | F: CAAGCAGCCCCCATATCTAA |
| R: CCAAGGGGATCCAGGATATT | |
| miR-3189-3p | F: ATGCTGCCCTTGGGTCTG |
| R: CACTTCCTCAGCACTTGTTGGTAT | |
| GAPDH | F: ATCATCAGCAATGCCTCC |
| R: CATCACGCCACAGTTTCC | |
| U6 | F: ATTGGAACGATACAGAGAAGATT |
| R: GGAACGCTTCACGAATTTG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bian, Y.; Guo, J.; Qiao, L.; Sun, X. miR-3189-3p Mimics Enhance the Effects of S100A4 siRNA on the Inhibition of Proliferation and Migration of Gastric Cancer Cells by Targeting CFL2. Int. J. Mol. Sci. 2018, 19, 236. https://doi.org/10.3390/ijms19010236
Bian Y, Guo J, Qiao L, Sun X. miR-3189-3p Mimics Enhance the Effects of S100A4 siRNA on the Inhibition of Proliferation and Migration of Gastric Cancer Cells by Targeting CFL2. International Journal of Molecular Sciences. 2018; 19(1):236. https://doi.org/10.3390/ijms19010236
Chicago/Turabian StyleBian, Yue, Junfu Guo, Linlin Qiao, and Xiuju Sun. 2018. "miR-3189-3p Mimics Enhance the Effects of S100A4 siRNA on the Inhibition of Proliferation and Migration of Gastric Cancer Cells by Targeting CFL2" International Journal of Molecular Sciences 19, no. 1: 236. https://doi.org/10.3390/ijms19010236




