Differences in Codon Usage Bias between Photosynthesis-Related Genes and Genetic System-Related Genes of Chloroplast Genomes in Cultivated and Wild Solanum Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Genomes and Coding Sequences
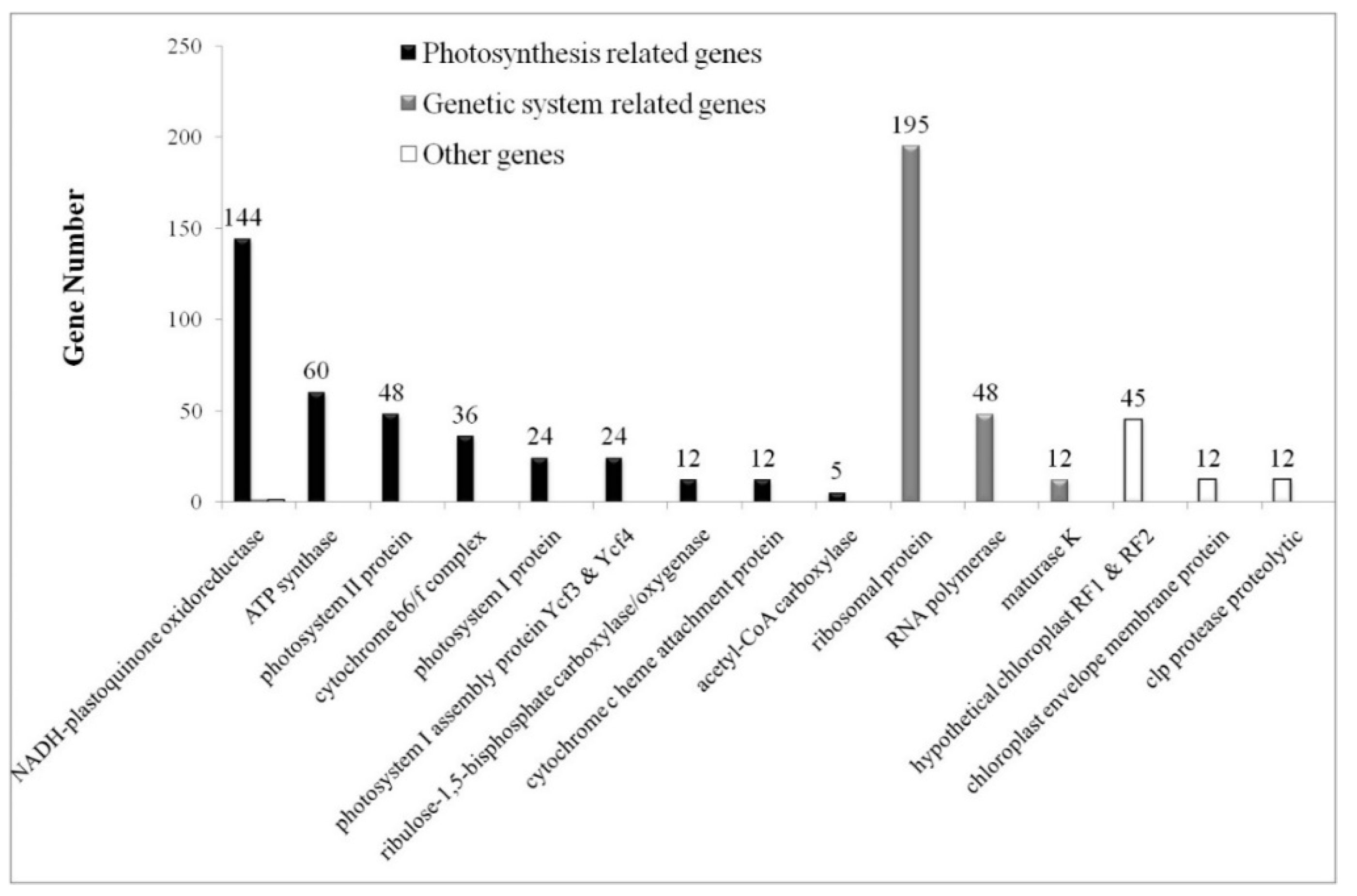
2.2. Classification of Photosynthesis Related and Genetic System Related Genes
2.3. Nucleotide Composition Analyses
2.4. Codon Usage Indices (CAI, ENc, GC3s, Gravy, and Aromo)
2.5. Relative Synonymous Codon Usage and Correspondence Analyses
3. Results
3.1. Codon Usage Indices
3.2. Codon Usage Biases
3.3. Parity Rule 2 Plot Analyses
3.4. Neutrality Plot Analyses
3.5. Correspondence Analyses
4. Discussion
4.1. Similarity in Codon Usage Among the Twelve Solanum Chloroplast Genomes
4.2. Impact of Selective Pressure on Shaping CUB in the Solanum Chloroplast Genomes
4.3. Different RSCU Values between Cultivated Species and Wild Species
4.4. Differences between Photo-Genes and Genet-Genes
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Frodin, D.G. History and concepts of big plant genera. Taxon 2004, 53, 753–776. [Google Scholar] [CrossRef]
- Weese, T.L.; Bohs, L. A three-gene phylogeny of the genus Solanum (Solanaceae). Syst. Bot. 2007, 32, 445–463. [Google Scholar] [CrossRef]
- Gargano, D.; Vezzi, A.; Scotti, N.; Gray, J.; Valle, G.; Grillo, S.; Cardi, T. The Complete Nucleotide Sequence Genome of Potato (Solanum tuberosum cv. Désirée) Chloroplast DNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/NC_008096 (accessed on 12 October 2018).
- Daniell, H.; Lee, S.-B.; Grevich, J.; Saski, C.; Quesada-Vargas, T.; Guda, C.; Tomkins, J.; Jansen, R.K. Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor. Appl. Genet. 2006, 112, 1503–1518. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z. The completed eight chloroplast genomes of tomato from Solanum genus. Mit DNA 2016, 27, 4155–4157. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.S.; Cheon, K.S.; Hong, S.Y.; Cho, J.H.; Im, J.S.; Mekapogu, M.; Yu, Y.S.; Park, T.H. Complete chloroplast genome sequences of Solanum commersonii and its application to chloroplast genotype in somatic hybrids with Solanum tuberosum. Plant Cell Rep. 2016, 35, 2113–2123. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.S.; Park, T.H. Complete chloroplast genome sequence of Solanum nigrum and development of markers for the discrimination of S. nigrum. Hort. Environ. Biotechnol. 2016, 57, 69–78. [Google Scholar] [CrossRef]
- Grantham, R.; Gautier, C.; Gouy, M. Codon frequencies in 119 individual genes confirm corsistent choices of degenerate bases according to genome type. Nucleic Acids Res. 1980, 8, 1893–1912. [Google Scholar] [CrossRef] [PubMed]
- Osawa, S.; Ohama, T.; Yamao, F.; Muto, A.; Jukes, T.H.; Ozeki, H.; Umesono, K. Directional mutation pressure and transfer RNA in choice of the third nucleotide of synonymous two-codon sets. Proc. Natl. Acad. Sci. USA 1988, 85, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Mackiewicz, P.; Gierlik, A.; Kowalczuk, M.; Dudek, M.R.; Cebrat, S. How does replication-associated mutational pressure influence amino acid composition of proteins? Gen. Res. 1999, 9, 409–416. [Google Scholar]
- Mackiewicz, P.; Kowalczuk, M.; Mackiewicz, D.; Nowicka, A.; Dudkiewicz, M.; Łaszkiewicz, A.; Dudek, M.R.; Cebrat, S. Replication associated mutational pressure generating long-range correlation in DNA. Phys. A 2002, 314, 646–654. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.-H. Codon usage in regulatory genes in Escherichia coli does not reflect selection for “rare” codons. Nucleic Acids Res. 1986, 14, 7737–7749. [Google Scholar] [CrossRef] [PubMed]
- Akashi, H. Synonymous codon usage in Drosophila melanogaster: Natural selection and translational accuracy. Genetics 1994, 136, 927–935. [Google Scholar] [PubMed]
- Ikemura, T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: A proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 1981, 151, 389–409. [Google Scholar] [CrossRef]
- Kanaya, S.; Yamada, Y.; Kudo, Y.; Ikemura, T. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: Gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 1999, 238, 143–155. [Google Scholar] [CrossRef]
- Chiapello, H.; Lisacek, F.; Caboche, M.; Hénaut, A. Codon usage and gene function are related in sequences of Arabidopsis thaliana. Gene 1998, 209, 1–38. [Google Scholar] [CrossRef]
- Moriyama, E.N.; Powell, J.R. Gene length and codon usage bias in Drosophila melanogaster, Saccharomyces cerevisiae and Escherichia coli. Nucleic Acids Res. 1998, 26, 3188–3193. [Google Scholar] [CrossRef] [PubMed]
- Duret, L.; Mouchiroud, D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. USA 1999, 96, 4482–4487. [Google Scholar] [CrossRef] [PubMed]
- Orešič, M.; Shalloway, D. Specific correlations between relative synonymous codon usage and protein secondary structure. J. Mol. Biol. 1998, 281, 31–48. [Google Scholar] [CrossRef] [PubMed]
- Romero, H.; Zavala, A.; Musto, H. Codon usage in Chlamydia trachomatis is the result of strand-specific mutational biases and a complex pattern of selective forces. Nucleic Acids Res. 2000, 28, 2084–2090. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Weems, M.; Wilke, C.O. Translationally optimal codons associate with structurally sensitive sites in proteins. Mol. Biol. Evol. 2009, 26, 1571–1580. [Google Scholar] [CrossRef] [PubMed]
- Sau, K.; Deb, A. Temperature influences synonymous codon and amino acid usage biases in the phages infecting extremely thermophilic prokaryotes. In Silico Biol. 2009, 9, 1–9. [Google Scholar] [PubMed]
- Błażej, P.; Mackiewicz, D.; Wnętrzak, M.; Mackiewicz, P. The impact of selection at the amino acid level on the usage of synonymous codons. G3 Genes Genet. 2017, 7, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Lee, W.; Hottes, A.K.; Shapiro, L.; Mcadams, H.H. Codon usage between genomes is constrained by genome-wide mutational processes. Proc. Natl. Acad. Sci. USA 2004, 101, 3480–3485. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, A.; Saxonov, S.; Gilbert, W. Regularities of context-dependent codon bias in eukaryotic genes. Nucleic Acids Res. 2002, 30, 1192–1197. [Google Scholar] [CrossRef] [PubMed]
- Morton, B.R. The role of context-dependent mutations in generating compositional and codon usage bias in grass chloroplast DNA. J. Mol. Evol. 2003, 56, 616–629. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Morton, B.R. Codon adaptation of plastid genes. PLoS ONE 2016, 11, e0154306. [Google Scholar] [CrossRef] [PubMed]
- Shokri, E. Codon bias patterns in photosynthetic genes of halophytic grass Aeluropus littoralis. J. Plant Mol. Breed. 2014, 3, 12–20. [Google Scholar]
- Chen, X.; Cai, X.; Chen, Q.; Zhou, H.; Cai, Y.; Ben, A. Factors affecting synonymous codon usage bias in chloroplast genome of Oncidium Gower Ramsey. Evol. Bioinform. 2011, 7, 271–278. [Google Scholar]
- Leister, D. Chloroplast research in the genomic age. Trends Genet. 2003, 19, 47–56. [Google Scholar] [CrossRef]
- Rosenberg, M.S.; Subramanian, S.; Kumar, S. Patterns of transitional mutation biases within and among mammalian genomes. Mol. Biol. Evol. 2003, 20, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Willey, D.L.; Gray, J.C. An open reading frame encoding a putative haem-binding polypeptide is cotranscribed with the pea chloroplast gene for apocytochrome f. Plant Mol. Biol. 1990, 15, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Maurizi, M.R.; Clark, W.P.; Katayama, Y.; Rudikoff, S.; Pumphrey, J.; Bowers, B.; Gottesman, S. Sequence and structure of Clp P, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J. Biol. Chem. 1990, 265, 12536–12545. [Google Scholar] [PubMed]
- Sueoka, N. Directional mutation pressure and neutral molecular evolution. Proc. Natl. Acad. Sci. USA 1988, 85, 2653–2657. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q. Analysis of codon usage pattern in the radioresistant bacterium Deinococcus radiodurans. Biosystems 2006, 85, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Greenacre, M.J. Theory and Applications of Correspondence Analysis; Academic Press: London, UK, 1984. [Google Scholar]
- Wright, F. The ‘effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef]
- Sueoka, N. Translation-coupled violation of parity rule 2 in human genes is not the cause of heterogeneity of the DNA G+C content of third codon position. Gene 1999, 238, 53–58. [Google Scholar] [CrossRef]
- Sueoka, N. Intrastrand parity rules of DNA base composition and usage biases of synonymous codons. J. Mol. Evol. 1995, 40, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M.; Cowe, E.; Higgins, D.G.; Shields, D.C.; Wolfe, K.H.; Wright, F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988, 16, 8207–8211. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D. Plastid chromosomes: Structure and evolution. In The Molecular Biology of Plastids; Springer: Vienna, Austria, 1991; Volume 7A, pp. 5–53. [Google Scholar]
- Raubeson, L.A.; Jansen, R.K. Chloroplast genomes of plants. In Diversity and Evolution of Plants-Genotypic and Phenotypic Variation in Higher Plants; CABI Publishing: Wallingford, UK, 2005; pp. 45–68. [Google Scholar]
- Ravi, V.; Khurana, J.P.; Tyagi, A.K.; Khurana, P. An update on chloroplast genomes. Plant Syst. Evol. 2008, 271, 101–122. [Google Scholar] [CrossRef]
- Hershberg, R.; Petrov, D.A. General rules for optimal codon choice. PLoS Genet. 2009, 5, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Sharp, P.M. Identification of functional open reading frames in chloroplast genomes. Gene 1988, 66, 215–222. [Google Scholar] [CrossRef]
- Morton, B.R. Chloroplast DNA codon use: Evidence for selection at the psbA locus based on tRNA availability. J. Mol. Evol. 1993, 37, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Long, W.; Li, X. Patterns of synonymous codon usage bias in chloroplast genomes of seed plants. For. Stud. China 2008, 10, 235–242. [Google Scholar] [CrossRef]
- Hershberg, R.; Petrov, D.A. Evidence that mutation is universally biased towards at in bacteria. PLoS Genet. 2010, 6, e1001115. [Google Scholar] [CrossRef] [PubMed]
- Raven, J.A.; Allen, J.F. Genomics and chloroplast evolution: What did cyanobacteria do for plants? Genome Biol. 2003, 4, 1–5. [Google Scholar] [CrossRef]
- Campbell, W.H.; Gowri, G. Codon usage in higher plants, green algae, and cyanobacteria. Plant. Physiol. 1990, 92, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kawabe, A.; Miyashita, N.T. Patterns of codon usage bias in three dicot and four monocot plant species. Genes Genet. Syst. 2003, 78, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Chargaff, E. Chemical specificity of nucleic acids and mechanism of their enzymatic degradation. Experientia 1950, 6, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Tillier, E.R.M.; Collins, R.A. The contributions of replication orientation, gene direction, and signal sequences to base-composition asymmetries in bacterial genomes. J. Mol. Evol. 2000, 50, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Charneski, C.A.; Honti, F.; Bryant, J.M.; Hurst, L.D.; Feil, E.J. Atypical at skew in firmicute genomes results from selection and not from mutation. PLoS Genet. 2011, 7, e1002283. [Google Scholar] [CrossRef] [PubMed]
- Necşulea, A.; Lobry, J.R. A new method for assessing the effect of replication on DNA base composition asymmetry. Mol. Biol. Evol. 2007, 24, 2169–2179. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Deng, P.; Feng, K.; Liu, P.; Du, X.; You, F.M.; Song, W. Comparative analysis of codon usage patterns in chloroplast genomes of the Asteraceae family. Plant. Mol. Biol. Rep. 2014, 32, 828–840. [Google Scholar] [CrossRef]
- Liu, Q.; Xue, Q. Comparative studies on codon usage pattern of chloroplasts and their host nuclear genes in four plant species. J. Genet. 2005, 84, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.R.; Nandhini, M.B.; Elango, M.; Kavitha, M.; Thilaga, S.; Sangeetha, N.; Rao, N.S.P.; Doss, G. Synonymous codon usage in chloroplast genome of Coffea arabica. Bioinformation 2012, 8, 1096–1104. [Google Scholar] [CrossRef] [PubMed]
- Park, T.H. The complete chloroplast genome of Solanum berthaultii, one of the potato wild relative species. Mit DNA Part B 2017, 2, 88–89. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.H. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Karlin, S.; Mrázek, J.; Campbell, A.; Kaiser, D. Characterizations of highly expressed genes of four fast-growing bacteria. J. Bacteriol. 2001, 183, 5025–5040. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Wang, R.; Zhang, C.; Wang, S.; He, W.; Zhang, J.; Liu, J.; Cai, Y.; Zhou, J.; Su, S. Genetic and evolutionary analysis of emerging H3N2 canine influenza virus. Emerg. Microbes Infect. 2018, 7, 73. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, X.-Q. Organellar genome copy number variation and integrity during moderate maturation of roots and leaves of maize seedlings. Curr. Genet. 2015, 61, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Q.; Chetrit, P.; Mathieu, C.; Vedel, F.; De Paepe, R.; Remy, R.; Ambard-Bretteville, F. Regeneration of cytoplasmic male sterile protoclones of Nicotiana sylvestris with mitochondrial variations. Curr. Genet. 1988, 13, 261–266. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, W.; Wu, Y.; Maliga, P.; Messing, J. RNA editing in chloroplasts of Spirodela polyrhiza, an aquatic monocotelydonous species. PLoS ONE 2015, 10, e0140285. [Google Scholar] [CrossRef] [PubMed]
- Lobry, J.R. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 1996, 13, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Nielsen, R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J. Mol. Evol. 1998, 46, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418. [Google Scholar] [PubMed]





| Black Nightshade | Potato | Tomato | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild Species | Cultivated Species | Wild Species | Cultivated Species | |||||||||
| S. nigrum | S. bulbocastanum | S. commersonii | S. tuberosum | S. neorickii | S. peruvianum | S. chilense | S. habrochaites | S. pimpinellifolium | S. galapagense | S. cheesmaniae | S. lycopersicum | |
| Accession No. | KM489055 | NC_007943 | KM489054 | NC_008096 | KP117025 | KP117026 | KP117021 | KP117023 | KP117027 | KP117022 | KP117020 | KP117024 |
| Genomic size (bp) | 155,432 | 155,371 | 155,525 | 155,296 | 155,513 | 155,561 | 155,528 | 155,465 | 155,442 | 155,474 | 155,520 | 155,452 |
| CDS | 86 | 85 | 86 | 84 | 83 | 83 | 83 | 83 | 83 | 83 | 83 | 83 |
| CDS>300bp | 58 | 58 | 58 | 58 | 57 | 57 | 57 | 57 | 57 | 57 | 57 | 57 |
| Lengthcds (aa) | 431.22 ± 475.46 | 426.60 ± 476.99 | 430.71 ± 475.30 | 426.38 ± 476.96 | 429.84 ± 479.54 | 429.96 ± 479.53 | 429.84 ± 479.54 | 429.84 ± 479.54 | 429.84 ± 479.54 | 429.84 ± 479.54 | 429.89 ± 479.54 | 429.76 ± 477.10 |
| GCgenome | 0.38 | 0.38 | 0.38 | 0.38 | 0.38 | 0.38 | 0.38 | 0.38 | 0.38 | 0.38 | 0.38 | 0.38 |
| GCcds | 0.39 ± 0.04 | 0.39 ± 0.04 | 0.39 ± 0.04 | 0.39 ± 0.04 | 0.39 ± 0.03 | 0.39 ± 0.03 | 0.39 ± 0.03 | 0.39 ± 0.04 | 0.39 ± 0.04 | 0.39 ± 0.03 | 0.39 ± 0.03 | 0.39 ± 0.04 |
| P1 | 0.49 ± 0.06 | 0.49 ± 0.06 | 0.49 ± 0.06 | 0.49 ± 0.06 | 0.49 ± 0.06 | 0.49 ± 0.06 | 0.49 ± 0.06 | 0.49 ± 0.06 | 0.49 ± 0.06 | 0.49 ± 0.06 | 0.49 ± 0.06 | 0.49 ± 0.06 |
| P2 | 0.40 ± 0.06 | 0.40 ± 0.06 | 0.40 ± 0.06 | 0.40 ± 0.06 | 0.40 ± 0.06 | 0.40 ± 0.06 | 0.40 ± 0.06 | 0.40 ± 0.06 | 0.40 ± 0.06 | 0.40 ± 0.06 | 0.40 ± 0.06 | 0.40 ± 0.06 |
| P12 | 0.44 ± 0.05 | 0.45 ± 0.05 | 0.44 ± 0.05 | 0.45 ± 0.05 | 0.45 ± 0.05 | 0.45 ± 0.05 | 0.45 ± 0.05 | 0.45 ± 0.05 | 0.45 ± 0.05 | 0.45 ± 0.05 | 0.45 ± 0.05 | 0.45 ± 0.05 |
| P3 | 0.27 ± 0.04 | 0.27 ± 0.04 | 0.27 ± 0.04 | 0.27 ± 0.04 | 0.27 ± 0.04 | 0.27 ± 0.04 | 0.27 ± 0.04 | 0.27 ± 0.04 | 0.27 ± 0.04 | 0.27 ± 0.04 | 0.27 ± 0.04 | 0.27 ± 0.04 |
| CAI (ref. psbA) | 0.21 ± 0.04 | 0.21 ± 0.04 | 0.21 ± 0.04 | 0.21 ± 0.04 | 0.21 ± 0.04 | 0.21 ± 0.04 | 0.21 ± 0.04 | 0.21 ± 0.04 | 0.21 ± 0.04 | 0.21 ± 0.04 | 0.21 ± 0.04 | 0.21 ± 0.04 |
| CAI (ref. E. coli) | 0.17 ± 0.04 | 0.17 ± 0.04 | 0.17 ± 0.04 | 0.17 ± 0.04 | 0.17 ± 0.04 | 0.17 ± 0.04 | 0.17 ± 0.04 | 0.17 ± 0.04 | 0.17 ± 0.04 | 0.17 ± 0.04 | 0.17 ± 0.04 | 0.17 ± 0.04 |
| ENc | 48.54 ± 4.44 | 48.44 ± 4.23 | 48.63 ± 4.18 | 48.46 ± 4.14 | 48.69 ± 4.32 | 48.66 ± 4.31 | 48.65 ± 4.30 | 48.69 ± 4.29 | 48.70 ± 4.35 | 48.70 ± 4.36 | 48.70 ± 4.36 | 48.56 ± 4.23 |
| Gravy | −0.06 ± 0.50 | −0.06 ± 0.50 | −0.05 ± 0.50 | −0.06 ± 0.50 | −0.05 ± 0.50 | −0.05 ± 0.50 | −0.05 ± 0.50 | −0.05 ± 0.50 | −0.05 ± 0.50 | −0.05 ± 0.50 | −0.05 ± 0.50 | −0.06 ± 0.50 |
| Aromo | 0.10 ± 0.04 | 0.10 ± 0.04 | 0.10 ± 0.04 | 0.10 ± 0.04 | 0.10 ± 0.04 | 0.10 ± 0.04 | 0.10 ± 0.04 | 0.10 ± 0.04 | 0.10 ± 0.04 | 0.10 ± 0.04 | 0.10 ± 0.04 | 0.10 ± 0.04 |
| Photo-Genes | Genet-Genes | Other Genes | |
|---|---|---|---|
| CDS | 581 | 353 | 75 |
| CDS>300bp | 365 | 255 | 69 |
| Lengthcds (aa) | 355.23 ± 187.59 | 314.09 ± 331.29 | 1246.13 ± 960.28 |
| GCcds | 0.39 ± 0.03 | 0.39 ± 0.03 | 0.37 ± 0.03 |
| P1 | 0.50 ± 0.06 | 0.48 ± 0.06 | 0.46 ± 0.07 |
| P2 | 0.39 ± 0.04 | 0.43 ± 0.07 | 0.33 ± 0.03 |
| P12 | 0.45 ± 0.04 | 0.46 ± 0.05 | 0.39 ± 0.05 |
| P3 | 0.27 ± 0.03 | 0.26 ± 0.04 | 0.31 ± 0.04 |
| CAI (ref. psbA) | 0.23 ± 0.05 | 0.19 ± 0.03 | 0.20 ± 0.03 |
| CAI (ref. E. coli) | 0.18 ± 0.04 | 0.15 ± 0.03 | 0.17 ± 0.02 |
| ENc | 48.65 ± 4.08 | 47.57 ± 4.30 | 52.31 ± 2.75 |
| Gravy | 0.26 ± 0.44 | −0.46 ± 0.21 | −0.20 ± 0.33 |
| Aromo | 0.12 ± 0.04 | 0.07 ± 0.03 | 0.12 ± 0.01 |
| Amino Acid | Codon | Black Nightshade | Potato | Tomato | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild Species | Cultivated Species | Wild Species | Cultivated Species | ||||||||||
| S. nigrum | S. bulbocastanum | S. commersonii | S. tuberosum | S. neorickii | S. peruvianum | S. chilense | S. habrochaites | S. pimpinellifolium | S. galapagense | S. cheesmaniae | S. lycopersicum | ||
| Phe | UUU | 1.00 | 1.78 | 1.00 | 1.00 | 1.78 | 1.78 | 1.78 | 1.78 | 1.78 | 1.78 | 1.78 | 1.78 |
| UUC | 1.00 | 0.22 | 1.00 | 1.00 | 0.22 | 0.22 | 0.22 | 0.22 | 0.22 | 0.22 | 0.22 | 0.22 | |
| Leu | UUA | 0.33 | 2.50@ | 0.35 | 0.35 | 2.50@ | 2.50@ | 2.50@ | 2.50@ | 2.50@ | 2.50@ | 2.50@ | 0.75 |
| UUG | 2.00 | 1.50 | 2.12 | 2.12 | 1.50 | 1.50 | 1.50 | 1.50 | 1.50 | 1.50 | 1.50 | 2.75 | |
| CUU | 2.33 | 1.00 | 2.12@ | 2.12 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.50 | |
| CUC | 0.00- | 0.25 | 0.00- | 0.00- | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | |
| CUA | 1.33 | 0.50 | 1.41 | 1.41 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | |
| CUG | 0.00- | 0.25 | 0.00- | 0.00- | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | |
| Ile | AUU | 1.31 | 1.66 | 1.41 | 1.41 | 1.61 | 1.61 | 1.61 | 1.61 | 1.61 | 1.61 | 1.61 | 1.58 |
| AUC | 0.00- | 0.41 | 0.00- | 0.00- | 0.43 | 0.43 | 0.43 | 0.43 | 0.43 | 0.43 | 0.43 | 0.79 | |
| AUA | 1.69 | 0.93 | 1.59 | 1.59@ | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 | 0.63 | |
| Met | AUG | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Val | GUU | 2.50 | 1.33 | 2.50@ | 2.50 | 1.71 | 1.71 | 1.71 | 1.71 | 1.71 | 1.71 | 1.71 | 3.11 * |
| GUC | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | |
| GUA | 1.50 | 0.67 | 1.50 | 1.50 | 0.57 | 0.57 | 0.57 | 0.57 | 0.57 | 0.57 | 0.57 | 0.89 | |
| GUG | 0.00- | 2.00@ | 0.00- | 0.00- | 1.71@ | 1.71@ | 1.71@ | 1.71@ | 1.71@ | 1.71@ | 1.71@ | 0.00- | |
| Tyr | UAU | 2.00 | 2.00 | 2.00 | 2.00 | 1.67 | 1.67 | 1.67 | 1.67 | 1.67 | 1.67 | 1.67 | 1.67 |
| UAC | 0.00- | 0.00- | 0.00- | 0.00- | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | |
| STOP | UAA | 3.00 | 1.50 | 3.00 | 3.00 | 1.50 | 1.50 | 1.50 | 1.50 | 1.50 | 1.50 | 1.50 | 3.00 |
| UAG | 0.00- | 1.50 | 0.00- | 0.00- | 1.50 | 1.50 | 1.50 | 1.50 | 1.50 | 1.50 | 1.50 | 0.00- | |
| UGA | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | |
| His | CAU | 1.60 | 2.00 | 1.67 | 1.67 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 1.60 |
| CAC | 0.40 | 0.00- | 0.33 | 0.33 | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.40 | |
| Gln | CAA | 1.25 | 2.00 | 1.25 | 1.25 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 1.33 |
| CAG | 0.75 | 0.00- | 0.75 | 0.75 | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.67 | |
| Asn | AAU | 1.60 | 1.60 | 1.60 | 1.60 | 1.47 | 1.47 | 1.47 | 1.47 | 1.47 | 1.47 | 1.47 | 1.50 |
| AAC | 0.40 | 0.40 | 0.40 | 0.40 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.50 | |
| Lys | AAA | 1.47 | 1.73 | 1.47 | 1.47 | 1.73 | 1.73 | 1.73 | 1.73 | 1.73 | 1.73 | 1.73 | 1.17 |
| AAG | 0.53 | 0.27 | 0.53 | 0.53 | 0.27 | 0.27 | 0.27 | 0.27 | 0.27 | 0.27 | 0.27 | 0.83 | |
| Asp | GAU | 0.00- | 1.43@ | 0.00- | 0.00- | 1.43 | 1.43 | 1.43 | 1.43 | 1.43 | 1.43 | 1.43 | 0.80 |
| GAC | 2.00@ | 0.57 | 2.00 * | 2.00 * | 0.57 | 0.57 | 0.57 | 0.57 | 0.57 | 0.57 | 0.57 | 1.20 * | |
| Glu | GAA | 1.82 | 1.17 | 1.82 | 1.82 | 1.17 | 1.17 | 1.17 | 1.17 | 1.17 | 1.17 | 1.17 | 1.75 |
| GAG | 0.18 | 0.83 | 0.18 | 0.18 | 0.83 | 0.83 | 0.83 | 0.83 | 0.83 | 0.83 | 0.83 | 0.25 | |
| Ser | UCU | 0.86 | 2.80 | 1.09 | 1.09 | 2.80 * | 2.80 * | 2.80 * | 2.80 * | 2.80 * | 2.80 * | 2.80 * | 0.29 |
| UCC | 1.43 | 0.40 | 1.36 | 1.36 | 0.40 | 0.40 | 0.40 | 0.40 | 0.40 | 0.40 | 0.40 | 0.86 | |
| UCA | 1.14 | 0.40 | 1.09 | 0.82 | 0.40 | 0.40 | 0.40 | 0.40 | 0.40 | 0.40 | 0.40 | 2.00 | |
| UCG | 0.86 | 0.80 | 0.82 | 1.09 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | 0.86 | |
| AGU | 1.43 | 1.60 | 1.36 | 1.36 | 1.60 | 1.60 | 1.60 | 1.60 | 1.60 | 1.60 | 1.60 | 1.71 | |
| AGC | 0.29 | 0.00- | 0.27 | 0.27 | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.29 | |
| Pro | CCU | 1.23 | 1.71 | 1.23 | 1.23 | 1.71 | 1.71 | 1.71 | 1.71 | 1.71 | 1.71 | 1.71 | 0.80 |
| CCC | 0.62 | 1.14 | 0.62 | 0.62 | 1.14 | 1.14 | 1.14 | 1.14 | 1.14 | 1.14 | 1.14 | 0.00- | |
| CCA | 1.23 | 0.00- | 1.23 | 1.23 | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 1.60 | |
| CCG | 0.92 | 1.14 | 0.92 | 0.92 | 1.14 | 1.14 | 1.14 | 1.14 | 1.14 | 1.14 | 1.14 | 1.60 | |
| Thr | ACU | 0.40 | 2.15@ | 0.40 | 0.40 | 2.15 | 2.15 | 2.15 | 2.15 | 2.15 | 2.15 | 2.15 | 0.67 |
| ACC | 0.80 | 1.54 | 0.80 | 0.80 | 1.54 | 1.54 | 1.54 | 1.54 | 1.54 | 1.54 | 1.54 | 0.67 | |
| ACA | 2.80 * | 0.31 | 2.80 * | 2.80 * | 0.31 | 0.31 | 0.31 | 0.31 | 0.31 | 0.31 | 0.31 | 2.67 * | |
| ACG | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | |
| Ala | GCU | 1.41 | 3.00@ | 1.18 | 1.18 | 3.00@ | 3.00@ | 3.00@ | 3.00@ | 3.00@ | 3.00@ | 3.00@ | 1.07 |
| GCC | 0.00- | 0.50 | 0.24 | 0.24 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 1.07 | |
| GCA | 2.59 * | 0.50 | 2.59 * | 2.59 * | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 1.87 * | |
| GCG | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | |
| Cys | UGU | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 |
| UGC | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | |
| Trp | UGG | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Arg | CGU | 1.59 | 0.63 | 1.45 | 1.45 | 0.63 | 0.63 | 0.63 | 0.63 | 0.63 | 0.63 | 0.63 | 1.80 |
| CGC | 0.18 | 0.00- | 0.18 | 0.18 | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.30 | |
| CGA | 1.41 | 3.16@ | 1.64 | 1.64 | 3.16@ | 3.16@ | 3.16@ | 3.16@ | 3.16@ | 3.16@ | 3.16@ | 1.20 | |
| CGG | 0.35 | 0.00- | 0.36 | 0.36 | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.00- | 0.30 | |
| AGA | 1.76 | 1.42 | 1.45 | 1.45 | 1.42 | 1.42 | 1.42 | 1.42 | 1.42 | 1.42 | 1.42 | 1.20 | |
| AGG | 0.71 | 0.79 | 0.91 | 0.91 | 0.79 | 0.79 | 0.79 | 0.79 | 0.79 | 0.79 | 0.79 | 1.20 | |
| Gly | GGU | 1.11 | 1.85 | 1.11 | 1.11 | 1.85 | 1.85 | 1.85 | 1.85 | 1.85 | 1.85 | 1.85 | 0.44 |
| GGC | 0.44 | 0.31 | 0.67 | 0.67 | 0.31 | 0.31 | 0.31 | 0.31 | 0.31 | 0.31 | 0.31 | 0.89 | |
| GGA | 2.22 | 1.23 | 2.00 | 2.00 | 1.23 | 1.23 | 1.23 | 1.23 | 1.23 | 1.23 | 1.23 | 2.67 | |
| GGG | 0.22 | 0.62 | 0.22 | 0.22 | 0.62 | 0.62 | 0.62 | 0.62 | 0.62 | 0.62 | 0.62 | 0.00- | |
| Photo-Genes | Genet-Genes | Other Genes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Axis1 | Axis2 | Axis3 | Axis4 | Axis1 | Axis2 | Axis3 | Axis4 | Axis1 | Axis2 | Axis3 | Axis4 | |
| GCcds | 0.699 * | 0.238 * | 0.154 * | 0.171 * | 0.173 * | −0.058 | 0.083 | 0.026 | −0.346 * | −0.099 | −0.724 * | 0.280 @ |
| GC3s | 0.629 * | −0.127 @ | 0.151 * | 0.083 | −0.030 | −0.589 * | −0.288 * | 0.010 | 0.419 * | 0.644 * | −0.760 * | 0.133 |
| Lengthcds | 0.170 * | 0.169 * | 0.294 * | 0.167 * | 0.047 | −0.058 | −0.563 * | −0.094 | 0.404 * | 0.720 * | −0.418 * | −0.524 * |
| ENc | 0.254 * | −0.401 * | 0.001 | −0.143 * | −0.154 @ | −0.151 @ | −0.520 * | −0.066 | 0.314 * | 0.450 * | −0.163 | 0.547 * |
| CAI | 0.191 * | 0.545 * | −0.043 | −0.206 * | 0.262 * | 0.459 * | −0.235 * | 0.287 * | −0.037 | −0.609 * | 0.791 * | −0.022 |
| Gravy | −0.193 * | 0.154 * | 0.182 * | −0.193 * | 0.016 | −0.111 | −0.211 * | 0.043 | 0.091 | −0.450 * | −0.024 | 0.771 * |
| Aromo | 0.179 * | −0.226 * | 0.449 * | −0.201 * | 0.112 | −0.111 | −0.248 * | −0.238 * | 0.485 * | 0.158 | 0.559 * | 0.091 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.; Zhang, L.; Wang, W.; Zhang, Z.; Du, H.; Qu, Z.; Li, X.-Q.; Xiang, H. Differences in Codon Usage Bias between Photosynthesis-Related Genes and Genetic System-Related Genes of Chloroplast Genomes in Cultivated and Wild Solanum Species. Int. J. Mol. Sci. 2018, 19, 3142. https://doi.org/10.3390/ijms19103142
Zhang R, Zhang L, Wang W, Zhang Z, Du H, Qu Z, Li X-Q, Xiang H. Differences in Codon Usage Bias between Photosynthesis-Related Genes and Genetic System-Related Genes of Chloroplast Genomes in Cultivated and Wild Solanum Species. International Journal of Molecular Sciences. 2018; 19(10):3142. https://doi.org/10.3390/ijms19103142
Chicago/Turabian StyleZhang, Ruizhi, Li Zhang, Wei Wang, Zhu Zhang, Huihui Du, Zheng Qu, Xiu-Qing Li, and Heng Xiang. 2018. "Differences in Codon Usage Bias between Photosynthesis-Related Genes and Genetic System-Related Genes of Chloroplast Genomes in Cultivated and Wild Solanum Species" International Journal of Molecular Sciences 19, no. 10: 3142. https://doi.org/10.3390/ijms19103142
APA StyleZhang, R., Zhang, L., Wang, W., Zhang, Z., Du, H., Qu, Z., Li, X.-Q., & Xiang, H. (2018). Differences in Codon Usage Bias between Photosynthesis-Related Genes and Genetic System-Related Genes of Chloroplast Genomes in Cultivated and Wild Solanum Species. International Journal of Molecular Sciences, 19(10), 3142. https://doi.org/10.3390/ijms19103142





