Exploring Protein–Protein Interaction in the Study of Hormone-Dependent Cancers
Abstract
:1. Introduction
2. Co-Immunoprecipitation
3. Bioluminescence Resonance Energy Transfer and Förster Resonance Energy Transfer
4. Protein Detection Methods
5. Proximity Ligation Assay
6. Super-Resolution Microscopy
7. Future Perspectives on Exploring PPI in Hormone-Dependent Cancers
Author Contributions
Funding
Conflicts of Interest
References
- Kumar, S.; Nussinov, R. Close-range electrostatic interactions in proteins. ChemBioChem 2002, 3, 604–617. [Google Scholar] [CrossRef]
- Lowenstein, E.J.; Daly, R.J.; Batzer, A.G.; Li, W.; Margolis, B.; Lammers, R.; Ullrich, A.; Skolnik, E.Y.; Bar-Sagi, D.; Schlessinger, J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell 1992, 70, 431–442. [Google Scholar] [CrossRef]
- Rozakis-Adcock, M.; McGlade, J.; Mbamalu, G.; Pelicci, G.; Daly, R.; Li, W.; Batzer, A.; Thomas, S.; Brugge, J.; Pelicci, P.G.; et al. Association of the Shc and Grb2/Sem5 SH2-containing proteins is implicated in activation of the Ras pathway by tyrosine kinases. Nature 1992, 360, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Khvorova, A.; Marshall, W.; Sorkin, A. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J. Biol. Chem. 2004, 279, 16657–16661. [Google Scholar] [CrossRef] [PubMed]
- Jorissen, R.N.; Walker, F.; Pouliot, N.; Garrett, T.P.; Ward, C.W.; Burgess, A.W. Epidermal growth factor receptor: Mechanisms of activation and signalling. Exp. Cell Res. 2003, 284, 31–53. [Google Scholar] [CrossRef]
- Sasano, H.; Harada, N. Intratumoral aromatase in human breast, endometrial, and ovarian malignancies. Endocr. Rev. 1998, 19, 593–607. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Utsunomiya, H.; Yaegashi, N.; Sasano, H. Biological roles of estrogen and progesterone in human endometrial carcinoma—New developments in potential endocrine therapy for endometrial cancer. Endocr. J. 2007, 54, 667–679. [Google Scholar] [CrossRef] [PubMed]
- McInerney, E.M.; Katzenellenbogen, B.S. Different regions in activation function-1 of the human estrogen receptor required for antiestrogen- and estradiol-dependent transcription activation. J. Biol. Chem. 1996, 271, 24172–24178. [Google Scholar] [CrossRef] [PubMed]
- Delaunay, F.; Pettersson, K.; Tujague, M.; Gustafsson, J.A. Functional differences between the amino-terminal domains of estrogen receptors α and β. Mol. Pharmacol. 2000, 58, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Metivier, R.; Penot, G.; Flouriot, G.; Pakdel, F. Synergism between ERα transactivation function 1 (AF-1) and AF-2 mediated by steroid receptor coactivator protein-1: Requirement for the AF-1 α-helical core and for a direct interaction between the N- and C-terminal domains. Mol. Endocrinol. 2001, 15, 1953–1970. [Google Scholar] [CrossRef] [PubMed]
- Beato, M. Gene regulation by steroid hormones. Cell 1989, 56, 335–344. [Google Scholar] [CrossRef]
- Leygue, E.; Dotzlaw, H.; Watson, P.H.; Murphy, L.C. Expression of estrogen receptor β1, β2, and β5 messenger RNAs in human breast tissue. Cancer Res. 1999, 59, 1175–1179. [Google Scholar] [PubMed]
- Cavallini, A.; Messa, C.; Pricci, M.; Caruso, M.L.; Barone, M.; Di Leo, A. Distribution of estrogen receptor subtypes, expression of their variant forms, and clinicopathological characteristics of human colorectal cancer. Dig. Dis. Sci. 2002, 47, 2720–2728. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, X.; Shen, P.; Loggie, B.W.; Chang, Y.; Deuel, T.F. Identification, cloning, and expression of human estrogen receptor-α36, a novel variant of human estrogen receptor-α66. Biochem. Biophys. Res. Commun. 2005, 336, 1023–1027. [Google Scholar] [CrossRef] [PubMed]
- Rao, J.; Jiang, X.; Wang, Y.; Chen, B. Advances in the understanding of the structure and function of ER-α36,a novel variant of human estrogen receptor-α. J. Steroid Biochem. Mol. Biol. 2011, 127, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Cowley, S.M.; Hoare, S.; Mosselman, S.; Parker, M.G. Estrogen receptors α and β form heterodimers on DNA. J. Biol. Chem. 1997, 272, 19858–19862. [Google Scholar] [CrossRef] [PubMed]
- Tamrazi, A.; Carlson, K.E.; Daniels, J.R.; Hurth, K.M.; Katzenellenbogen, J.A. Estrogen receptor dimerization: Ligand binding regulates dimer affinity and dimer dissociation rate. Mol. Endocrinol. 2002, 16, 2706–2719. [Google Scholar] [CrossRef] [PubMed]
- Groner, A.C.; Brown, M. Role of steroid receptor and coregulator mutations in hormone-dependent cancers. J. Clin. Investig. 2017, 127, 1126–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Cavailles, V.; Dauvois, S.; L’Horset, F.; Lopez, G.; Hoare, S.; Kushner, P.J.; Parker, M.G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995, 14, 3741–3751. [Google Scholar] [CrossRef] [PubMed]
- Lavinsky, R.M.; Jepsen, K.; Heinzel, T.; Torchia, J.; Mullen, T.M.; Schiff, R.; Del-Rio, A.L.; Ricote, M.; Ngo, S.; Gemsch, J.; et al. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc. Natl. Acad. Sci. USA 1998, 95, 2920–2925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansson, L.; Thomsen, J.S.; Damdimopoulos, A.E.; Spyrou, G.; Gustafsson, J.A.; Treuter, E. The orphan nuclear receptor SHP inhibits agonist-dependent transcriptional activity of estrogen receptor α and β. J. Biol. Chem. 1999, 274, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Yang, X.; Wang, Y.; Shen, X. In Vivo Analysis of Protein–Protein Interactions with Bioluminescence Resonance Energy Transfer (BRET): Progress and Prospects. Int. J. Mol. Sci. 2016, 17, 1704. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, N.; Terai, K.; Imanishi, A.; Kamioka, Y.; Sumiyama, K.; Jin, T.; Okada, Y.; Nagai, T.; Matsuda, M.A. platform of BRET-FRET hybrid biosensors for optogenetics, chemical screening, and in vivo imaging. Sci. Rep. 2018, 8, 8984. [Google Scholar] [CrossRef] [PubMed]
- Michnick, S.W.; Ear, P.H.; Landry, C.; Malleshaiah, M.K.; Messier, V. A toolkit of protein-fragment complementation assays for studying and dissecting large-scale and dynamic protein-protein interactions in living cells. Methods Enzymol. 2010, 470, 335–368. [Google Scholar] [PubMed]
- Yurlova, L.; Derks, M.; Buchfellner, A.; Hickson, I.; Janssen, M.; Morrison, D.; Stansfield, I.; Brown, C.J.; Ghadessy, F.J.; Lane, D.P.; et al. The fluorescent two-hybrid assay to screen for protein-protein interaction inhibitors in live cells: Targeting the interaction of p53 with Mdm2 and Mdm4. J. Biomol. Screen. 2014, 19, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Malleshaiah, M.; Tchekanda, E.; Michnick, S.W. Real-Time Protein-Fragment Complementation Assays for Studying Temporal, Spatial, and Spatiotemporal Dynamics of Protein-Protein Interactions in Living Cells. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ding, M.; Xue, B.; Hou, Y.; Sun, Y. Live Cell Visualization of Multiple Protein-Protein Interactions with BiFC Rainbow. ACS Chem. Biol. 2018, 13, 1180–1188. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.; Baek, J.; Han, S.B.; Cho, S. Facile Analysis of Protein-Protein Interactions in Living Cells by Enriched Visualization of the P-body. BMB Rep. 2018, in press. [Google Scholar]
- Lin, T.; Scott, B.L.; Hoppe, A.D.; Chakravarty, S. FRETting About the Affinity of Bimolecular Protein-Protein Interactions. Protein Sci. 2018, in press. [Google Scholar] [CrossRef] [PubMed]
- Masters, S.C. Co-immunoprecipitation from transfected cells. Methods Mol. Biol. 2004, 261, 337–350. [Google Scholar] [PubMed]
- Tang, Z.; Takahashi, Y. Analysis of Protein-Protein Interaction by Co-IP in Human Cells. Methods Mol. Biol. 2018, 1794, 289–296. [Google Scholar] [PubMed]
- Mohammed, H.; D’Santos, C.; Serandour, A.A.; Ali, H.R.; Brown, G.D.; Atkins, A.; Rueda, O.M.; Holmes, K.A.; Theodorou, V.; Robinson, J.L.; et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013, 3, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Boute, N.; Jockers, R.; Issad, T. The use of resonance energy transfer in high-throughput screening: BRET versus FRET. Trends Pharmacol. Sci. 2002, 23, 351–354. [Google Scholar] [CrossRef]
- Lohse, M.J.; Bünemann, M.; Hoffmann, C.; Vilardaga, J.P.; Nikolaev, V.O. Monitoring receptor signaling by intramolecular FRET. Curr. Opin. Pharmacol. 2007, 7, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Förster, T. Energy transport and fluorescence. Naturwissenschafien 1946, 6, 166–175. (In German) [Google Scholar]
- Emmanouilidou, E.; Teschemacher, A.G.; Pouli, A.E.; Nicholls, L.I.; Seward, E.P.; Rutter, G.A. Imaging Ca2+ concentration changes at the secretory vesicle surface with a recombinant targeted cameleon. Curr. Biol. 1999, 9, 915–918. [Google Scholar] [CrossRef]
- Shinoda, H.; Shannon, M.; Nagai, T. Fluorescent Proteins for Investigating Biological Events in Acidic Environments. Int. J. Mol. Sci. 2018, 19, 1548. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Piston, D.W.; Johnson, C.H. A bioluminescence resonance energy transfer (BRET) system: Application to interacting circadian clock proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 151–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, A.A.; Hansen, J.L.; Sheikh, S.P.; Bräuner-Osborne, H. Probing intermolecular protein-protein interactions in the calcium-sensing receptor homodimer using bioluminescence resonance energy transfer (BRET). Eur. J. Biochem. 2002, 269, 5076–5087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eidne, K.A.; Kroeger, K.M.; Hanyaloglu, A.C. Applications of novel resonance energy transfer techniques to study dynamic hormone receptor interactions in living cells. Trends Endocrinol. Metab. 2002, 13, 415–421. [Google Scholar] [CrossRef]
- De, A.; Jasani, A.; Arora, R.; Gambhir, S.S. Evolution of BRET Biosensors from Live Cell to Tissue-Scale In Vivo Imaging. Front. Endocrinol. (Lausanne) 2013, 4, 131. [Google Scholar] [CrossRef] [PubMed]
- Dragulescu-Andrasi, A.; Chan, C.T.; De, A.; Massoud, T.F.; Gambhir, S.S. Bioluminescence resonance energy transfer (BRET) imaging of protein-protein interactions within deep tissues of living subjects. Proc. Natl. Acad. Sci. USA 2011, 108, 12060–12065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tung, J.K.; Berglund, K.; Gutekunst, C.A.; Hochgeschwender, U.; Gross, R.E. Bioluminescence imaging in live cells and animals. Neurophotonics 2016, 3, 025001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michelini, E.; Mirasoli, M.; Karp, M.; Virta, M.; Roda, A. Development of a bioluminescence resonance energy-transfer assay for estrogen-like compound in vivo monitoring. Anal. Chem. 2004, 76, 7069–7076. [Google Scholar] [CrossRef] [PubMed]
- Yasgar, A.; Jadhav, A.; Simeonov, A.; Coussens, N.P. AlphaScreen-Based Assays: Ultra-High-Throughput Screening for Small-Molecule Inhibitors of Challenging Enzymes and Protein-Protein Interactions. Methods Mol. Biol. 2016, 1439, 77–98. [Google Scholar] [PubMed]
- Rouleau, N.; Turcotte, S.; Mondou, M.H.; Roby, P.; Bossé, R. Development of a versatile platform for nuclear receptor screening using AlphaScreen. J. Biomol. Screen. 2003, 8, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Yalow, R.S.; Berson, S.A. Immunoassay of endogenous plasma insulin in man. Clin. Investig. 1960, 39, 1157–1175. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, J.; Rippe, D.F.; Panfili, P.R. Comparison of enzymelinked immunosorhent assay and radioimmunoassay for prostatespecific acid phosphatase in prostatic disease. Clin. Chem. 1982, 28, 183–186. [Google Scholar] [PubMed]
- Holt, J.A.; Bolanos, J. Enzyme-linked immunochemical measurement of estrogen receptor in gynecologic tumors, and an overview of steroid receptors in ovarian carcinoma. Clin. Chem. 1986, 32, 1836–1843. [Google Scholar] [PubMed]
- Porstmann, T.; Kiessig, S.T. Enzyme immunoassay techniques. An overview. J. Immunol. Methods 1992, 150, 5–21. [Google Scholar] [CrossRef]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed]
- Burnette, W.N. “Western Blotting”: Electrophoretic Transfer of Proteins from Sodium Dodecyl Sulfate-Polyacrylamide Gels to Unmodified Nitrocellulose and Radiographic Detection with Antibody and Radioiodinated Protein A. Anal. Biochem. 1981, 112, 195–203. [Google Scholar] [CrossRef]
- Peluso, P.; Wilson, D.S.; Do, D.; Tran, H.; Venkatasubbaiah, M.; Quincy, D.; Heidecker, B.; Poindexter, K.; Tolani, N.; Phelan, M.; et al. Optimizing antibody immobilization strategies for the construction of protein microarrays. Anal. Biochem. 2003, 312, 113–124. [Google Scholar] [CrossRef]
- Watanabe, M.; Guo, W.; Zou, S.; Sugiyo, S.; Dubner, R.; Ren, K. Antibody array analysis of peripheral and blood cytokine levels in rats after masseter inflammation. Neurosci. Lett. 2005, 382, 128–133. [Google Scholar] [CrossRef] [PubMed]
- de Jager, W.; te Velthuis, H.; Prakken, B.J.; Kuis, W.; Rijkers, G.T. Simultaneous detection of 15 human cytokines in a single sample of stimulated peripheral blood mononuclear cells. Clin. Diagn. Lab. Immunol. 2003, 10, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Coons, A.H.; Creech, H.J.; Jones, R.N. Immunological properties of an antibody containing a fluorescent group. Proc. Soc. Exp. Biol. Med. 1941, 47, 200–202. [Google Scholar] [CrossRef]
- Jacobs, T.W.; Gown, A.M.; Yaziji, H.; Barnes, M.J.; Schnitt, S.J. Specificity of HercepTest in determining HER-2/neu status of breast cancers using the United States Food and Drug Administration-approved scoring system. J. Clin. Oncol. 1999, 17, 1983–1987. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.R. Predictive Biomarkers and Companion Diagnostics. The Future of Immunohistochemistry—‘In situ proteomics’, or just a ‘stain’? Appl. Immunohistochem. Mol. Morphol. 2014, 22, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Erskine, L.P.; Mary, L.M. Chapter 1: Immune complexing. In Immuno-Gold Electron Microscopy in Virus Diagnosis and Research; Hyatt, A.D., Eaton, B.T., Eds.; CRC Press: Boca Raton, FL, USA, 1993; pp. 3–24. ISBN 0-8493-675-X. [Google Scholar]
- Bridoux, F.; Leung, N.; Hutchison, C.A.; Touchard, G.; Sethi, S.; Fermand, J.P.; Picken, M.M.; Herrera, G.A.; Kastritis, E.; Merlini, G.; et al. Diagnosis of monoclonal gammopathy of renal significance. Kidney Int. 2015, 87, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Figueres, M.L.; Beaume, J.; Vuiblet, V.; Rabant, M.; Bassilios, N.; Herody, M.; Touchard, G.; Noël, L.H. Crystalline light chain proximal tubulopathy with chronic renal failure and silicone gel breast implants: 1 case report. Hum. Pathol. 2015, 46, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, A.; De Wever, O. Rab27 GTPases Distribute Extracellular Nanomaps for Invasive Growth and Metastasis: Implications for Prognosis and Treatment. Int. J. Mol. Sci. 2013, 14, 9883–9892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kocanova, S.; Mazaheri, M.; Caze-Subra, S.; Bystricky, K. Ligands specify estrogen receptor α nuclear localization and degradation. BMC Cell Biol. 2010, 11, 98. [Google Scholar] [CrossRef] [PubMed]
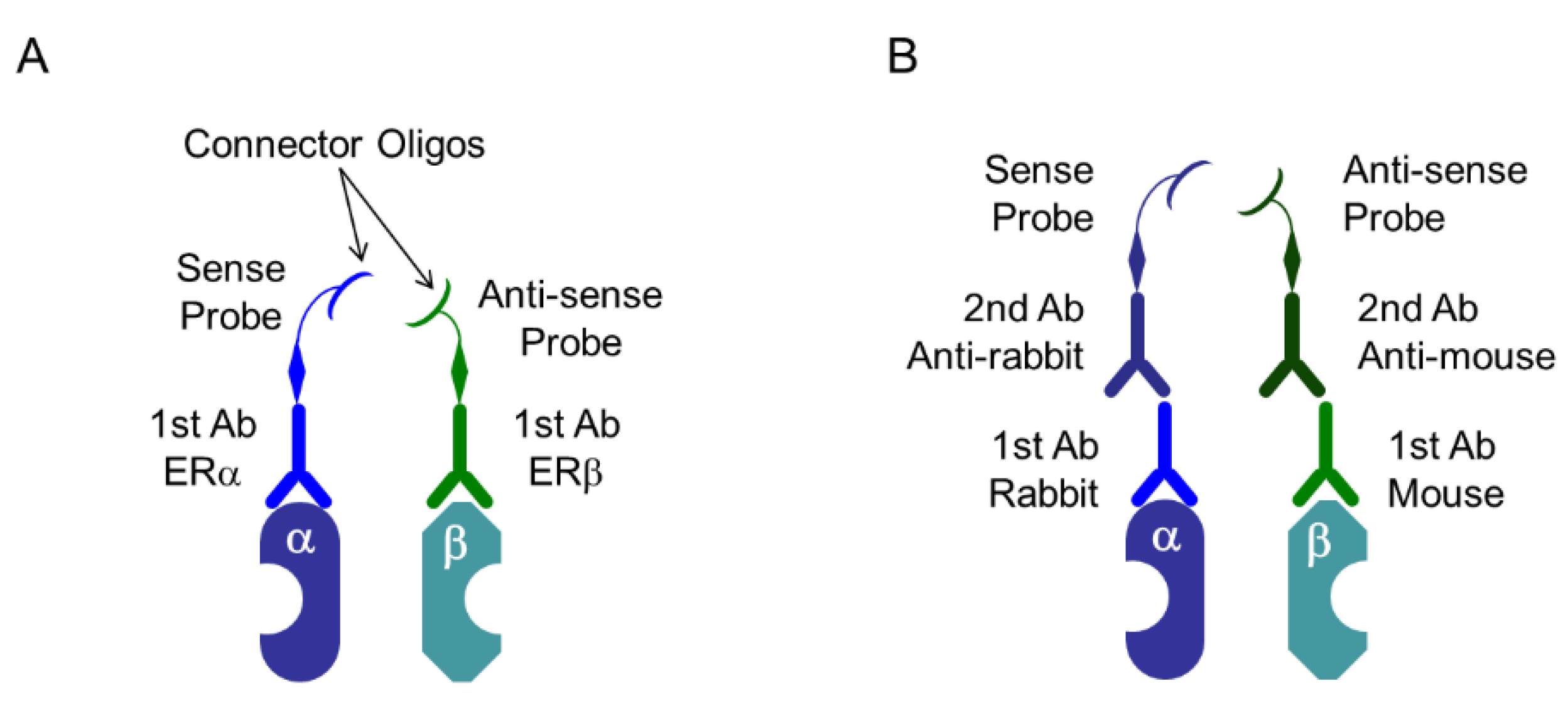
- Söderberg, O.; Gullberg, M.; Jarvius, M.; Ridderstrale, K.; Leuchowius, K.J.; Jarvius, J.; Wester, K.; Hydbring, P.; Bahram, F.; Larsson, L.G.; et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 2006, 3, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Söderberg, O.; Leuchowius, K.J.; Gullberg, M.; Jarvius, M.; Weibrecht, I.; Larsson, L.G.; Landegren, U. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods 2008, 45, 227–232. [Google Scholar] [CrossRef] [PubMed]
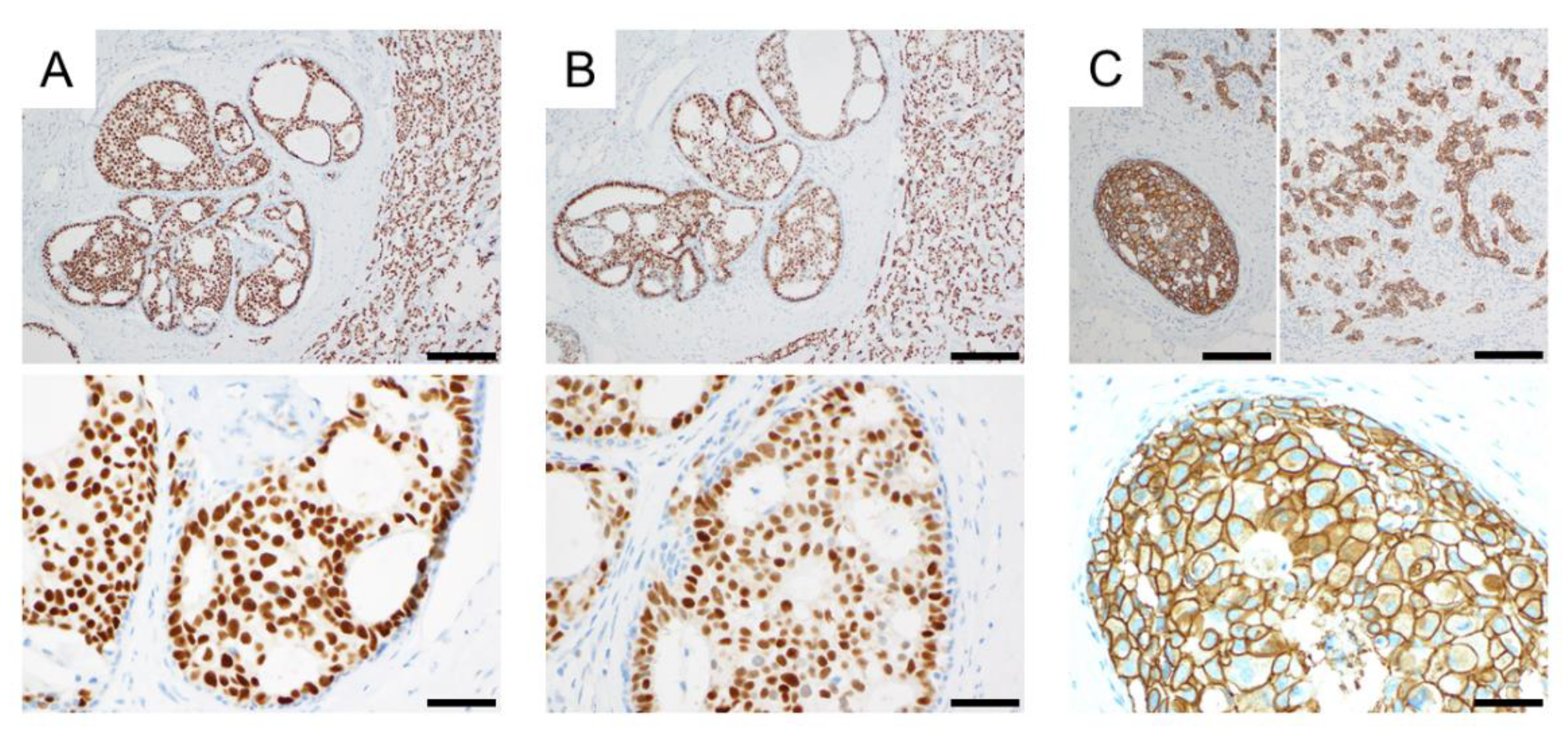
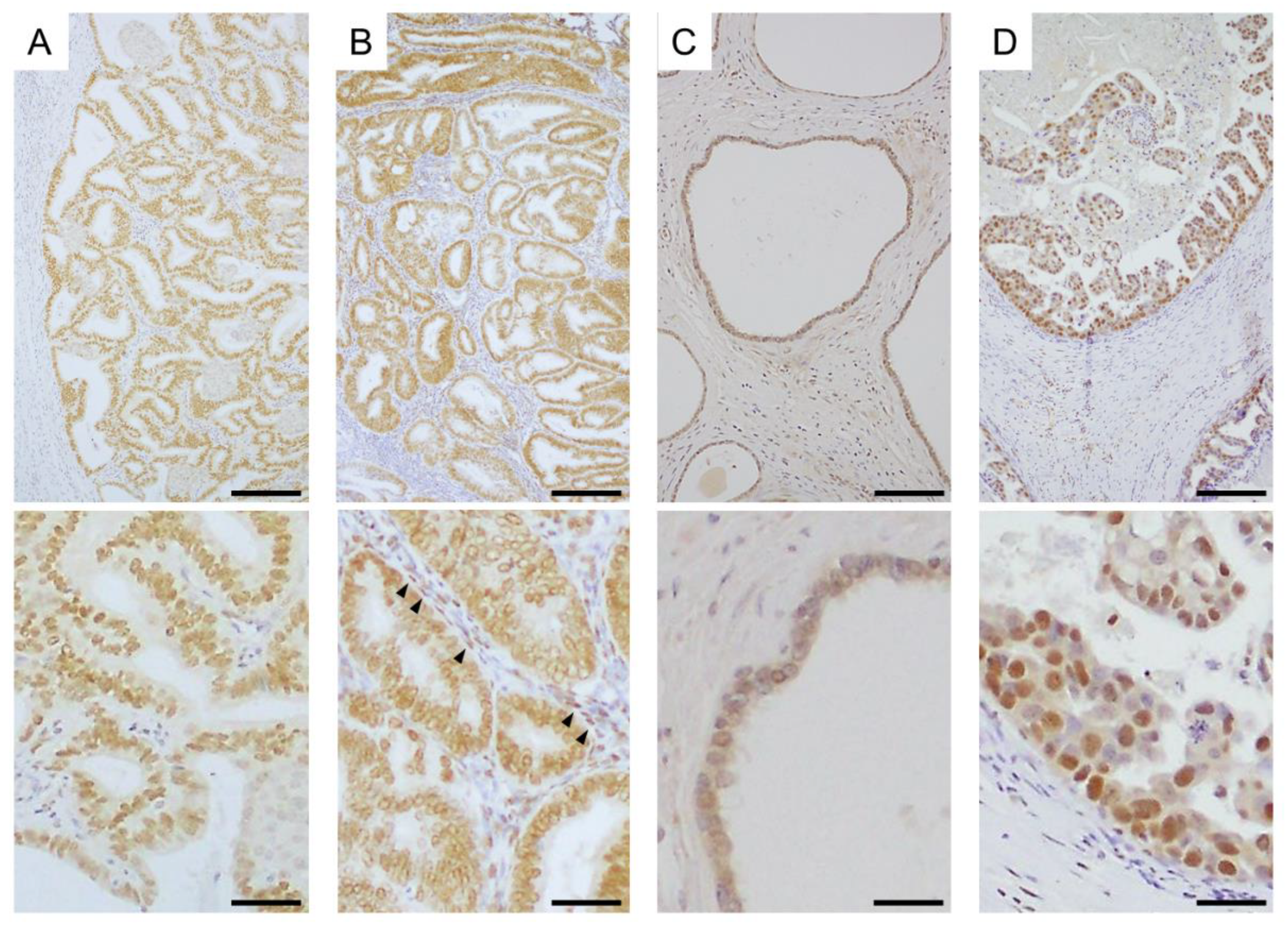
- Iwabuchi, E.; Miki, Y.; Ono, K.; Onodera, Y.; Suzuki, T.; Hirakawa, H.; Ishida, T.; Ohuchi, N.; Sasano, H. In situ detection of estrogen receptor dimers in breast carcinoma cells in archival materials using proximity ligation assay (PLA). J. Steroid Biochem. Mol. Biol. 2017, 165, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Iwabuchi, E.; Miki, Y.; Ono, K.; Onodera, Y.; Sasano, H. In Situ Evaluation of Estrogen Receptor Dimers in Breast Carcinoma Cells: Visualization of Protein-Protein Interactions. Acta Histochem. Cytochem. 2017, 50, 85–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, S.; Inoue, S.; Watanabe, T.; Orimo, A.; Hosoi, T.; Ouchi, Y.; Muramatsu, M. Molecular cloning and characterization of human estrogen receptor βcx: A potential inhibitor ofestrogen action in human. Nucleic Acids Res. 1998, 26, 3505–3512. [Google Scholar] [CrossRef] [PubMed]
- Omoto, Y.; Eguchi, H.; Yamamoto-Yamaguchi, Y.; Hayashi, S. Estrogen receptor (ER) β1 and ERβcx/β2 inhibit ERα function differently in breast cancer cell line MCF7. Oncogene 2003, 22, 5011–5020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, Y.K.; Mak, P.; Hassan, S.; Ho, S.M. Estrogen receptor (ER)-β isoforms: A key to understanding ER-β signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 13162–13167. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Lu, B.; Leygue, E.; Murphy, L.C. Putative functional characteristics of human estrogen receptor-β isoforms. J. Mol. Endocrinol. 2003, 30, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Flanders, K.C.; Heger, C.D.; Conway, C.; Tang, B.; Sato, M.; Dengler, S.L.; Goldsmith, P.K.; Hewitt, S.M.; Wakefield, L.M. Brightfield proximity ligation assay reveals both canonical and mixed transforming growth factor-β/bone morphogenetic protein Smad signaling complexes in tissue sections. J. Histochem. Cytochem. 2014, 62, 846–863. [Google Scholar] [CrossRef] [PubMed]
- Zieba, A.; Wählby, C.; Hjelm, F.; Jordan, L.; Berg, J.; Landegren, U.; Pardali, K. Bright-field microscopy visualization of proteins and protein complexes by in situ proximity ligation with peroxidase detection. Clin. Chem. 2010, 56, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Mehraein-Ghomi, F.; Kegel, S.J.; Church, D.R.; Schmidt, J.S.; Reuter, Q.R.; Saphner, E.L.; Basu, H.S.; Wilding, G. Targeting androgen receptor and JunD interaction for prevention of prostate cancer progression. Prostate 2014, 74, 792–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwabuchi, E.; Miki, Y.; Kanai, A.; Miyashita, M.; Kijima, G.; Hirakawa, H.; Suzuki, T.; Ishida, T.; Sasano, H. The interaction between carcinoembryonic antigen-related cell adhesion molecule 6 and HER2 is associated with therapeutic efficacy of trastuzumab in breast cancer. J. Pathol. 2018, in press. [Google Scholar] [CrossRef] [PubMed]
- Kanthala, S.; Banappagari, S.; Gokhale, A.; Liu, Y.Y.; Xin, G.; Zhao, Y.; Jois, S. Novel Peptidomimetics for Inhibition of HER2:HER3 Heterodimerization I HER2-Positive Breast Cancer. Chem. Biol. Drug Des. 2015, 85, 702–714. [Google Scholar] [CrossRef] [PubMed]
- Falkenberg, N.; Anastasov, N.; Höfig, I.; Bashkueva, K.; Lindner, K.; Höfler, H.; Rosemann, M.; Aubele, M. Additive impact of HER2-/PTK6-RNAi on interactions with HER3 or IGF-1R leads to reduced breast cancer progression in vivo. Mol. Oncol. 2015, 9, 282–294. [Google Scholar] [CrossRef] [PubMed]
- Barros, F.F.; Abdel-Fatah, T.M.; Moseley, P.; Nolan, C.C.; Durham, A.C.; Rakha, E.A.; Chan, S.; Ellis, I.O.; Green, A.R. Characterisation of HER heterodimers in breast cancer using in situ proximity ligation assay. Breast Cancer Res. Treat. 2014, 144, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Spears, M.; Taylor, K.J.; Munro, A.F.; Cunningham, C.A.; Mallon, E.A.; Twelves, C.J.; Cameron, D.A.; Thomas, J.; Bartlett, J.M. In situ detection of HER2:HER2 and HER2:HER3 protein-protein interactions demonstrates prognostic significance in early breast cancer. Breast Cancer Res. Treat. 2012, 132, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Ellebaek, S.; Brix, S.; Grandal, M.; Lantto, J.; Horak, I.D.; Kragh, M.; Poulsen, T.T. Pan-HER-An antibody mixture targeting EGFR, HER2 and HER3 abrogates preformed and ligand-induced EGFR homo- and heterodimers. Int. J. Cancer 2016, 139, 2095–2105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahjat, M.; Bloedjes, T.A.; van der Veen, A.; de Wilde, G.; Maas, C.; Guikema, J.E.J. Detection and Visualization of DNA Damage-induced Protein Complexes in Suspension Cell Cultures Using the Proximity Ligation Assay. J. Vis. Exp. 2017, 124, e55703. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson, S.; Horecka, J.; Brustugun, O.T.; Schlingemann, J.; Koong, A.C.; Tibshirani, R.; Davis, R.W. Multiplexed proximity ligation assays to profile putative plasma biomarkers relevant to pancreatic and ovarian cancer. Clin. Chem. 2008, 54, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Leuchowius, K.J.; Weibrecht, I.; Landegren, U.; Gedda, L.; Soderberg, O. Flow cytometric in situ proximity ligation analyses of proteininteractions and post-translational modification of the epidermal growth factor receptor family. Cytom. A 2009, 75, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Blokzijl, A.; Friedman, M.; Pontén, F.; Landegren, U. Profiling protein expression and interactions: Proximity ligation as a tool for personalized medicine. J. Intern. Med. 2010, 268, 232–245. [Google Scholar] [CrossRef] [PubMed]
- Ward, E.N.; Pal, R. Image scanning microscopy: An overview. J. Microsc. 2017, 266, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Vangindertael, J.; Camacho, R.; Sempels, W.; Mizuno, H.; Dedecker, P.; Janssen, K.P.F. An introduction to optical super-resolution microscopy for the adventurous biologist. Methods Appl. Fluoresc. 2018, 6, 022003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Htun, H.; Holth, L.T.; Walker, D.; Davie, J.R.; Hager, G.L. Direct visualization of the human estrogen receptor α reveals a role for ligand in the nuclear distribution of the receptor. Mol. Biol. Cell 1999, 10, 471–486. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Goto, K.; Saitoh, M.; Yanase, T.; Nomura, M.; Okabe, T.; Takayanagi, R.; Nawata, H. Activation function-1 domain of androgen receptor contributes to the interaction between subnuclear splicing factor compartment and nuclear receptor compartment. Identification of the p102 U5 small nuclear ribonucleoprotein particle-binding protein as a coactivator for the receptor. J. Biol. Chem. 2002, 277, 30031–30039. [Google Scholar] [PubMed]
- Ochiai, I.; Matsuda, K.; Nishi, M.; Ozawa, H.; Kawata, M. Imaging analysis of subcellular correlation of androgen receptor and estrogen receptor α in single living cells using green fluorescent protein color variants. Mol. Endocrinol. 2004, 18, 26–42. [Google Scholar] [CrossRef] [PubMed]
- Sharp, Z.D.; Mancini, M.G.; Hinojos, C.A.; Dai, F.; Berno, V.; Szafran, A.T.; Smith, K.P.; Lele, T.P.; Ingber, D.E.; Mancini, M.A. Estrogen-receptor-α exchange and chromatin dynamics are ligand- and domain-dependent. J. Cell Sci. 2006, 119, 4101–4116, Erratum in 2006, 119, 4365. [Google Scholar] [CrossRef] [PubMed]
- Thorley, J.A.; Pike, J.; Rappoport, J.Z. Chapter 14: Super-resolution Microscopy: A Comparison of Commercially Available Options. In Fluorescence Microscopy, Super-Resolution and Other Novel Techniques; Cornea, A., Conn, P.M., Eds.; Elsevier Academic Press: Amsterdam, The Netherlands, 2014; pp. 199–212. ISBN 978-0-12-409513-7. [Google Scholar]
- Habuchi, S. Super-resolution molecular and functional imaging of nanoscale architectures in life and materials science. Front. Bioeng. Biotechnol. 2014, 2, 20. [Google Scholar] [CrossRef] [PubMed]
- Jonas, K.C.; Fanelli, F.; Huhtaniemi, I.T.; Hanyaloglu, A.C. Single molecule analysis of functionally asymmetric G protein-coupled receptor (GPCR) oligomers reveals diverse spatial and structural assemblies. J. Biol. Chem. 2015, 290, 3875–3892. [Google Scholar] [CrossRef] [PubMed]
- Jonas, K.C.; Chen, S.; Virta, M.; Mora, J.; Franks, S.; Huhtaniemi, I.; Hanyaloglu, A.C. Temporal reprogramming of calcium signalling via crosstalk of gonadotrophin receptors that associate as functionally asymmetric heteromers. Sci. Rep. 2018, 8, 2239. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, R.; Müller, P.; Hildenbrand, G.; Hausmann, M.; Cremer, C. Analysis of Her2/neu membrane protein clusters in different types of breast cancer cells using localization microscopy. J. Microsc. 2011, 242, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Hausmann, M.; Ilić, N.; Pilarczyk, G.; Lee, J.H.; Logeswaran, A.; Borroni, A.P.; Krufczik, M.; Theda, F.; Waltrich, N.; Bestvater, F.; et al. Challenges for Super-Resolution Localization Microscopy and Biomolecular Fluorescent Nano-Probing in Cancer Research. Int. J. Mol. Sci. 2017, 18, 2066. [Google Scholar] [CrossRef] [PubMed]
- Borroto-Escuela, D.O.; Romero-Fernandez, W.; Garriga, P.; Ciruela, F.; Narvaez, M.; Tarakanov, A.O.; Palkovits, M.; Agnati, L.F.; Fuxe, K. G protein-coupled receptor heterodimerization in the brain. Methods Enzymol. 2013, 521, 281–294. [Google Scholar] [PubMed]
- Jeanquartier, F.; Jean-Quartier, C.; Holzinger, A. Integrated web visualization for protein-protein interaction databases. BMC Bioinform. 2015, 16, 195. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.L.; Tamrazi, A.; Collins, M.L.; Katzenellenbogen, J.A. Design, synthesis, and in vitro biological evaluation of small molecule inhibitors of estrogen receptor α coactivator binding. J. Med. Chem. 2004, 47, 600–611. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Moore, T.W.; Gunther, J.R.; Kim, M.S.; Rhoden, E.; Du, Y.; Fu, H.; Snyder, J.P.; Katzenellenbogen, J.A. Discovering small-molecule estrogen receptor α/coactivator binding inhibitors: High-throughput screening, ligand development, and models for enhanced potency. ChemMedChem 2011, 6, 654–666. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, G.; Gallo, D.; Cossy, J.; Laïos, I.; Larsimont, D.; Laurent, G.; Jacquot, Y. Peptides targeting estrogen receptor α-potential applications for breast cancer treatment. Curr. Pharm. Des. 2011, 17, 2632–2653. [Google Scholar] [CrossRef] [PubMed]
- Lempereur, M.; Majewska, C.; Brunquers, A.; Wongpramud, S.; Valet, B.; Janssens, P.; Dillemans, M.; Van Nedervelde, L.; Gallo, D. Tetrahydro-iso-α Acids Antagonize Estrogen Receptor Alpha Activity in MCF-7 Breast Cancer Cells. Int. J. Endocrinol. 2016, 2016, 9747863. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Munuganti, R.S.N.; Lallous, N.; Dalal, K.; Yoon, J.S.; Sharma, A.; Yamazaki, T.; Cherkasov, A.; Rennie, P.S. Benzothiophenone Derivatives Targeting Mutant Forms of Estrogen Receptor-α in Hormone-Resistant Breast Cancers. Int. J. Mol. Sci. 2018, 19, 579. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Gustafsson, J.Å. Estrogen receptor mutations and functional consequences for breast cancer. Trends Endocrinol. Metab. 2015, 26, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Gimzewski, J.K.; Joachim, C. Nanoscale science of single molecules using local probes. Science 1999, 283, 1683–1688. [Google Scholar] [CrossRef] [PubMed]
- Sitterberg, J.; Ozcetin, A.; Ehrhardt, C.; Bakowsky, U. Utilising atomic force microscopy for the characterisation of nanoscale drug delivery systems. Eur. J. Pharm. Biopharm. 2010, 74, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Berthier, A.; Elie-Caille, C.; Lesniewska, E.; Delage-Mourroux, R.; Boireau, W. Label-free sensing and atomic force spectroscopy for the characterization of protein-DNA and protein-protein interactions: Application to estrogen receptors. J. Mol. Recognit. 2011, 24, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, G.; Laïos, I.; Elie-Caille, C.; Leiber, D.; Laurent, G.; Lesniewska, E.; Tanfin, Z.; Jacquot, Y. ERα dimerization: A key factor for the weak estrogenic activity of an ERα modulator unable to compete with estradiol in binding assays. J. Recept. Signal Transduct. Res. 2017, 37, 149–166. [Google Scholar] [CrossRef] [PubMed]
- Jacquot, Y.; Spaggiari, D.; Schappler, J.; Lesniewska, E.; Rudaz, S.; Leclercq, G. ERE-dependent transcription and cell proliferation: Independency of these two processes mediated by the introduction of a sulfone function into the weak estrogen estrothiazine. Eur. J. Pharm. Sci. 2017, 109, 169–181. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miki, Y.; Iwabuchi, E.; Ono, K.; Sasano, H.; Ito, K. Exploring Protein–Protein Interaction in the Study of Hormone-Dependent Cancers. Int. J. Mol. Sci. 2018, 19, 3173. https://doi.org/10.3390/ijms19103173
Miki Y, Iwabuchi E, Ono K, Sasano H, Ito K. Exploring Protein–Protein Interaction in the Study of Hormone-Dependent Cancers. International Journal of Molecular Sciences. 2018; 19(10):3173. https://doi.org/10.3390/ijms19103173
Chicago/Turabian StyleMiki, Yasuhiro, Erina Iwabuchi, Katsuhiko Ono, Hironobu Sasano, and Kiyoshi Ito. 2018. "Exploring Protein–Protein Interaction in the Study of Hormone-Dependent Cancers" International Journal of Molecular Sciences 19, no. 10: 3173. https://doi.org/10.3390/ijms19103173




