Genome-Wide Association Study Reveals Novel Genomic Regions Associated with 10 Grain Minerals in Synthetic Hexaploid Wheat
Abstract
:1. Introduction
2. Results and Discussion
2.1. Phenotypic Variation for Grain Protein Content and Grain Minerals
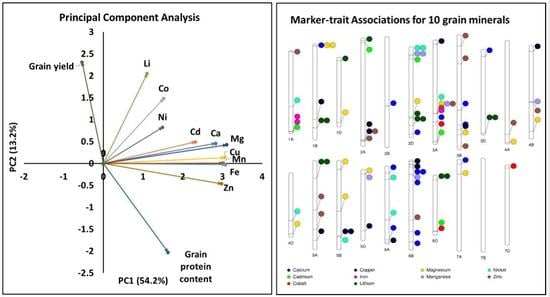
2.2. Principal Component Analysis and Phenotypic Correlation
2.3. Selection of Top-Ranking Genotypes
2.4. Population Structure and Genome-Wide Association Study
2.4.1. Calcium
2.4.2. Cadmium
2.4.3. Cobalt, Lithium, and Nickel
2.4.4. Copper
2.4.5. Iron
2.4.6. Magnesium
2.4.7. Manganese
2.4.8. Zinc
2.5. Relationship Between Grain Mineral Concentrations and Number of Favorable Alleles
2.6. Multi-Trait and Stable Marker-Trait Associations
2.7. Genes Underlying Marker-Trait Associations
3. Materials and Methods
3.1. Plant Materials and Experimental Design
3.2. Grain Yield, Thousand Kernel Weight, Grain Protein Concentration, and Grain Mineral Analysis
3.3. Phenotypic Data Analysis
3.4. Genotyping and SNP Discovery
3.5. Population Structure and Genome-Wide Association Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BLUP | Best Linear Unbiased Predictor |
| Ca | Calcium |
| Cd | Cadmium |
| Co | Cobalt |
| Cu | Copper |
| Fe | Iron |
| GBS | Genotyping-By-Sequencing |
| GPC | Grain Protein Concentration |
| GWAS | Genome-Wide Association Study |
| GY | Grain Yield |
| Li | Lithium |
| MAF | Minor Allele Frequency |
| Mg | Magnesium |
| Mn | Manganese |
| MTA | Marker-Trait Association |
| Ni | Nickel |
| PVE | Phenotypic Variance Explained |
| QTL | Quantitative Trait Loci |
| SHW | Synthetic Hexaploid Wheat |
| SNP | Single Nucleotide Polymorphism |
| TKW | Thousand Kernel Weight |
| Zn | Zinc |
References
- World Population Prospects. 2017. Available online: https://esa.un.org/unpd/wpp/Download/Standard/Population/ (accessed on 8 August 2018).
- Poudel, R.; Bhatta, M. Review of nutraceuticals and functional properties of whole wheat. J. Nutr. Food Sci. 2017, 7. [Google Scholar] [CrossRef]
- World food situation. Available online: http://www.fao.org/worldfoodsituation/csdb/en/ (accessed on 8 August 2018).
- Alomari, D.Z.; Eggert, K.; von Wirén, N.; Pillen, K.; Röder, M.S. Genome-wide association study of calcium accumulation in grains of european wheat cultivars. Front. Plant Sci. 2017, 8, 1797. [Google Scholar] [CrossRef] [PubMed]
- Velu, G.; Tutus, Y.; Gomez-Becerra, H.F.; Hao, Y.; Demir, L.; Kara, R.; Crespo-Herrera, L.A.; Orhan, S.; Yazici, A.; Singh, R.P.; et al. QTL mapping for grain zinc and iron concentrations and zinc efficiency in a tetraploid and hexaploid wheat mapping populations. Plant Soil 2017, 411, 81–99. [Google Scholar] [CrossRef]
- White, P.J.; Broadley, M.R. Biofortification of crops with seven mineral elements often lacking in human diets–iron, zinc, copper, calcium, magnesium, selenium and iodine. New Phytol. 2009, 182, 49–84. [Google Scholar] [CrossRef] [PubMed]
- Welch, R.M.; Graham, R.D. Breeding for micronutrients in staple food crops from a human nutrition perspective. J. Exp. Bot. 2004, 55, 353–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peleg, Z.; Cakmak, I.; Ozturk, L.; Yazici, A.; Jun, Y.; Budak, H.; Korol, A.B.; Fahima, T.; Saranga, Y. Quantitative trait loci conferring grain mineral nutrient concentrations in durum wheat × wild emmer wheat RIL population. Theor. Appl. Genet. 2009, 119, 353–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welch, R.M. Importance of seed mineral nutrient reserves in crop growth and development. In Mineral Nutrition of Crops: Fundamental Mechanisms and Implications; Rengel, Z., Ed.; CRC Press: Boca Raton, FL, USA, 1999; pp. 205–226. ISBN 978-1-56022-880-6. [Google Scholar]
- Shi, R.; Tong, Y.; Jing, R.; Zhang, F.; Zou, C. Characterization of quantitative trait loci for grain minerals in hexaploid wheat (Triticum aestivum L.). J. Integr. Agric. 2013, 12, 1512–1521. [Google Scholar] [CrossRef]
- Knox, R.E.; Pozniak, C.J.; Clarke, F.R.; Clarke, J.M.; Houshmand, S.; Singh, A.K. Chromosomal location of the cadmium uptake gene (Cdu1) in durum wheat. Genome 2009, 52, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Guttieri, M.J.; Baenziger, P.S.; Frels, K.; Carver, B.; Arnall, B.; Waters, B.M. Variation for Grain mineral concentration in a diversity panel of current and historical great plains hard winter wheat germplasm. Crop Sci. 2015, 55, 1035–1052. [Google Scholar] [CrossRef]
- Ozkan, H.; Brandolini, A.; Torun, A.; AltIntas, S.; Eker, S.; Kilian, B.; Braun, H.J.; Salamini, F.; Cakmak, I. Natural Variation and identification of microelements content in seeds of einkorn wheat (Triticum Monococcum). In Wheat Production in Stressed Environments; Developments in Plant Breeding; Springer: Dordrecht, The Netherlands, 2007; pp. 455–462. ISBN 978-1-4020-5496-9. [Google Scholar]
- Tiwari, V.K.; Rawat, N.; Chhuneja, P.; Neelam, K.; Aggarwal, R.; Randhawa, G.S.; Dhaliwal, H.S.; Keller, B.; Singh, K. Mapping of Quantitative trait loci for grain iron and zinc concentration in diploid A genome wheat. J. Hered. 2009, 100, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Genc, Y.; Verbyla, A.P.; Torun, A.A.; Cakmak, I.; Willsmore, K.; Wallwork, H.; McDonald, G.K. Quantitative trait loci analysis of zinc efficiency and grain zinc concentration in wheat using whole genome average interval mapping. Plant Soil 2009, 314, 49. [Google Scholar] [CrossRef]
- Krishnappa, G.; Singh, A.M.; Chaudhary, S.; Ahlawat, A.K.; Singh, S.K.; Shukla, R.B.; Jaiswal, J.P.; Singh, G.P.; Solanki, I.S. Molecular mapping of the grain iron and zinc concentration, protein content and thousand kernel weight in wheat (Triticum aestivum L.). PLoS ONE 2017, 12, e0174972. [Google Scholar] [CrossRef] [PubMed]
- Srinivasa, J.; Arun, B.; Mishra, V.K.; Singh, G.P.; Velu, G.; Babu, R.; Vasistha, N.K.; Joshi, A.K. Zinc and iron concentration QTL mapped in a Triticum spelta × T. aestivum cross. Theor. Appl. Genet. 2014, 127, 1643–1651. [Google Scholar] [CrossRef] [PubMed]
- Roshanzamir, H.; Kordenaeej, A.; Bostani, A. Mapping QTLs related to Zn and Fe concentrations in bread wheat (Triticum aestivum) grain using microsatellite markers. Iranian J. Genet. Plant Breed. 2013, 2, 10–17. [Google Scholar]
- Distelfeld, A.; Cakmak, I.; Peleg, Z.; Ozturk, L.; Yazici, A.M.; Budak, H.; Saranga, Y.; Fahima, T. Multiple QTL-effects of wheat Gpc-B1 locus on grain protein and micronutrient concentrations. Physiol. Plant. 2007, 129, 635–643. [Google Scholar] [CrossRef] [Green Version]
- Crespo-Herrera, L.A.; Velu, G.; Singh, R.P. Quantitative trait loci mapping reveals pleiotropic effect for grain iron and zinc concentrations in wheat. Ann. Appl. Biol. 2016, 169, 27–35. [Google Scholar] [CrossRef]
- Xu, Y.; An, D.; Liu, D.; Zhang, A.; Xu, H.; Li, B. Molecular mapping of QTLs for grain zinc, iron and protein concentration of wheat across two environments. Field Crops Res. 2012, 138, 57–62. [Google Scholar] [CrossRef]
- Cakmak, I. Enrichment of cereal grains with zinc: Agronomic or genetic biofortification? Plant Soil 2008, 302, 1–17. [Google Scholar] [CrossRef]
- Bhatta, M.; Morgounov, A.; Belamkar, V.; Poland, J.; Baenziger, P.S. Unlocking the novel genetic diversity and population structure of synthetic hexaploid wheat. BMC Genom. 2018, 19, 591. [Google Scholar] [CrossRef] [PubMed]
- Morgounov, A.; Abugalieva, A.; Akan, K.; Akın, B.; Baenziger, S.; Bhatta, M.; Dababat, A.A.; Demir, L.; Dutbayev, Y.; Bouhssini, M.E.; et al. High-yielding winter synthetic hexaploid wheats resistant to multiple diseases and pests. Plant Genet. Res. 2018, 16, 273–278. [Google Scholar] [CrossRef]
- Calderini, D.F.; Ortiz-Monasterio, I. Are synthetic hexaploids a means of increasing grain element concentrations in wheat? Euphytica 2003, 134, 169–178. [Google Scholar] [CrossRef]
- Bhatta, M.; Morgounov, A.; Belamkar, V.; Baenziger, P. Genome-Wide Association Study Reveals Novel Genomic Regions for Grain Yield and Yield-Related Traits in Drought-Stressed Synthetic Hexaploid Wheat. Int. J. Mol. Sci. 2018, 19, 3011. [Google Scholar] [CrossRef] [PubMed]
- Bhatta, M.; Regassa, T.; Rose, D.J.; Baenziger, P.S.; Eskridge, K.M.; Santra, D.K.; Poudel, R. Genotype, environment, seeding rate, and top-dressed nitrogen effects on end-use quality of modern Nebraska winter wheat. J. Sci. Food Agric. 2017, 97, 5311–5318. [Google Scholar] [CrossRef] [PubMed]
- Bhatta, M.; Eskridge, K.M.; Rose, D.J.; Santra, D.K.; Baenziger, P.S.; Regassa, T. Seeding rate, genotype, and topdressed nitrogen effects on yield and agronomic characteristics of winter wheat. Crop Sci. 2017, 57, 951–963. [Google Scholar] [CrossRef]
- Morgounov, A.; Gómez-Becerra, H.F.; Abugalieva, A.; Dzhunusova, M.; Yessimbekova, M.; Muminjanov, H.; Zelenskiy, Y.; Ozturk, L.; Cakmak, I. Iron and zinc grain density in common wheat grown in Central Asia. Euphytica 2007, 155, 193–203. [Google Scholar] [CrossRef] [Green Version]
- Graham, R.; Senadhira, D.; Beebe, S.; Iglesias, C.; Monasterio, I. Breeding for micronutrient density in edible portions of staple food crops: Conventional approaches. Field Crops Res. 1999, 60, 57–80. [Google Scholar] [CrossRef]
- Guttieri, M.J.; Baenziger, P.S.; Frels, K.; Carver, B.; Arnall, B.; Wang, S.; Akhunov, E.; Waters, B.M. Prospects for selecting wheat with increased zinc and decreased cadmium concentration in grain. Crop Sci. 2015, 55, 1712. [Google Scholar] [CrossRef]
- Moran Lauter, A.N.; Peiffer, G.A.; Yin, T.; Whitham, S.A.; Cook, D.; Shoemaker, R.C.; Graham, M.A. Identification of candidate genes involved in early iron deficiency chlorosis signaling in soybean (Glycine max) roots and leaves. BMC Genom. 2014, 15, 702. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Lan, P. Genome-wide analysis of overlapping genes regulated by iron deficiency and phosphate starvation reveals new interactions in Arabidopsis roots. BMC Res. Notes 2015, 8. [Google Scholar] [CrossRef] [PubMed]
- Laganowsky, A.; Gómez, S.M.; Whitelegge, J.P.; Nishio, J.N. Hydroponics on a chip: Analysis of the Fe deficient Arabidopsis thylakoid membrane proteome. J. Proteom. 2009, 72, 397–415. [Google Scholar] [CrossRef]
- Singh, S.P.; Jeet, R.; Kumar, J.; Shukla, V.; Srivastava, R.; Mantri, S.S.; Tuli, R. Comparative transcriptional profiling of two wheat genotypes, with contrasting levels of minerals in grains, shows expression differences during grain filling. PLoS ONE 2014, 9, e111718. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Huang, F.; Narsai, R.; Wu, J.; Giraud, E.; He, F.; Cheng, L.; Wang, F.; Wu, P.; Whelan, J.; et al. Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol. 2009, 151, 262–274. [Google Scholar] [CrossRef] [PubMed]
- King, K.E.; Peiffer, G.A.; Reddy, M.; Lauter, N.; Lin, S.F.; Cianzio, S.; Shoemaker, R.C. Mapping of iron and zinc quantitative trait loci in soybean for association to iron deficiency chlorosis resistance. J. Plant Nutr. 2013, 36, 2132–2153. [Google Scholar] [CrossRef]
- Elias, D.A.; Yang, F.; Mottaz, H.M.; Beliaev, A.S.; Lipton, M.S. Enrichment of functional redox reactive proteins and identification by mass spectrometry results in several terminal Fe(III)-reducing candidate proteins in Shewanella oneidensis MR-1. J. Microbiol. Methods 2007, 68, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Waters, B.M.; Uauy, C.; Dubcovsky, J.; Grusak, M.A. Wheat (Triticum aestivum) NAM proteins regulate the translocation of iron, zinc, and nitrogen compounds from vegetative tissues to grain. J. Exp. Bot. 2009, 60, 4263–4274. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.-W.; Huang, J.-H.; Li, C.-P.; Yang, L.-T.; Ye, X.; Lin, D.; Chen, L.-S. MicroRNA-mediated responses to long-term magnesium-deficiency in Citrus sinensis roots revealed by Illumina sequencing. BMC Genom. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Billard, V.; Maillard, A.; Garnica, M.; Cruz, F.; Garcia-Mina, J.-M.; Yvin, J.-C.; Ourry, A.; Etienne, P. Zn deficiency in Brassica napus induces Mo and Mn accumulation associated with chloroplast proteins variation without Zn remobilization. Plant Physiol. Biochem. 2015, 86, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Yuan, S.; Wen, X.; Xie, Z.; Lou, L.; Hu, B.; Cai, Q.; Xu, B. Transcriptome analysis of Cd-treated switchgrass root revealed novel transcripts and the importance of HSF/HSP network in switchgrass Cd tolerance. Plant Cell Rep. 2018. [Google Scholar] [CrossRef] [PubMed]
- Van De Mortel, J.E.; Schat, H.; Moerland, P.D.; Van Themaat, E.V.L.; Van Der Ent, S.; Blankestijn, H.; Ghandilyan, A.; Tsiatsiani, S.; Aarts, M.G.M. Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 2008, 31, 301–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Leonard, J.M.; von Zitzewitz, J.; James Peterson, C.; Ross, A.S.; Riera-Lizarazu, O. Marker–trait association analysis of kernel hardness and related agronomic traits in a core collection of wheat lines. Mol. Breed. 2014. [Google Scholar] [CrossRef]
- Waters, B.M.; Grusak, M.A. Quantitative trait locus mapping for seed mineral concentrations in two Arabidopsis thaliana recombinant inbred populations. New Phytol. 2008, 179, 1033–1047. [Google Scholar] [CrossRef] [PubMed]
- Chandran, D.; Sharopova, N.; Ivashuta, S.; Gantt, J.S.; VandenBosch, K.A.; Samac, D.A. Transcriptome profiling identified novel genes associated with aluminum toxicity, resistance and tolerance in Medicago truncatula. Planta 2008, 228, 151–166. [Google Scholar] [CrossRef] [PubMed]
- SAS 9.4 Product Documentation. Available online: https://support.sas.com/documentation/94/ (accessed on 16 August 2018).
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 16 August 2018).
- Poland, J.A.; Brown, P.J.; Sorrells, M.E.; Jannink, J. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by- sequencing approach. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Glaubitz, J.C.; Casstevens, T.M.; Lu, F.; Harriman, J.; Elshire, R.J.; Sun, Q.; Buckler, E.S. TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLoS ONE 2014, 9, e90346. [Google Scholar] [CrossRef] [PubMed]
- International Wheat Genome Sequencing Consortium (IWGSC). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [PubMed]
- Belamkar, V.; Guttieri, M.J.; Hussain, W.; Jarquín, D.; El-basyoni, I.; Poland, J.; Lorenz, A.J.; Baenziger, P.S. Genomic selection in preliminary yield trials in a winter wheat breeding program. G3 2018, 8, 2735–2747. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef] [PubMed]



| Trait | 2016 | 2017 | Trials Combined | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Fold | Mean | Min | Max | Fold | Mean | Year (Yr) | Genotype (G) | G x Yr | H2 | |
| Ca (mg∙kg−1) | 47.5 | 167.2 | 3.5 | 103.1 | 21.6 | 84.5 | 3.9 | 44.3 | * | ** | *** | 0.41 |
| Cd (mg∙kg−1) | 0.03 | 0.1 | 3.44 | 0.07 | 0.02 | 0.13 | 7.68 | 0.07 | NS | * | NS | 0.28 |
| Co (mg∙kg−1) | 0.01 | 0.06 | 6.53 | 0.03 | 0.01 | 0.04 | 6.86 | 0.02 | *** | * | NS | 0.33 |
| Cu (mg∙kg−1) | 2.8 | 11.4 | 4.1 | 7.5 | 2.9 | 8.9 | 3.1 | 5.7 | NS | *** | *** | 0.63 |
| Fe (mg∙kg−1) | 17.7 | 61.8 | 3.5 | 40.2 | 15.4 | 67.7 | 4.4 | 38.5 | NS | *** | NS | 0.78 |
| Li (mg∙kg−1) | 0.04 | 0.23 | 6.43 | 0.09 | 0.13 | 1.07 | 8.43 | 0.52 | *** | * | NS | 0.35 |
| Mg (mg∙kg−1) | 617 | 2097 | 3 | 1391 | 659 | 2131 | 3 | 1458 | NS | *** | ** | 0.62 |
| Mn (mg∙kg−1) | 20.3 | 66.2 | 3.3 | 41.2 | 21.5 | 69.8 | 3.2 | 44.9 | NS | *** | ** | 0.67 |
| Ni (mg∙kg−1) | 0.21 | 2.22 | 10.81 | 0.91 | 0.13 | 1.16 | 8.75 | 0.48 | NS | *** | *** | 0.52 |
| Zn (mg∙kg−1) | 8.8 | 38.1 | 4.3 | 23.1 | 11.1 | 39.6 | 3.6 | 23 | NS | *** | NS | 0.65 |
| Grain protein (g∙kg−1) | 129.8 | 167.6 | 1.3 | 151.2 | 116.4 | 168.9 | 1.5 | 137.8 | *** | *** | ** | 0.68 |
| Grain yield (g∙m−2) | 54.3 | 530 | 9.8 | 259 | 194.7 | 479.5 | 2.5 | 290.1 | NS | * | * | 0.44 |
| Trait | Ca | Cd | Co | Cu | Fe | Li | Mg | Mn | Ni | Zn | GPC | GY | GYpTKW |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ca | 1 | 0.63 *** | 0.42 *** | 0.64 *** | 0.58 *** | 0.31 *** | 0.80 *** | 0.79 *** | 0.44 *** | 0.60 *** | 0.36 *** | −0.01 | −0.11 |
| Cd | 0.64 *** | 1 | 0.32 *** | 0.68 *** | 0.61 *** | 0.38 *** | 0.65 *** | 0.67 *** | 0.58 *** | 0.63 *** | 0.22 * | −0.03 | −0.40 *** |
| Co | 0.37 *** | 0.34 *** | 1 | 0.49 *** | 0.63 *** | 0.19 * | 0.49 *** | 0.49 *** | 0.54 *** | 0.53 *** | −0.05 | 0.13 | 0.01 |
| Cu | 0.82 *** | 0.67 *** | 0.42 *** | 1 | 0.79 *** | 0.27 ** | 0.84 *** | 0.81 *** | 0.43 *** | 0.89 *** | 0.22 * | 0.04 | −0.18 * |
| Fe | 0.80 *** | 0.67 *** | 0.46 *** | 0.89 *** | 1 | 0.24 ** | 0.79 *** | 0.82 *** | 0.48 *** | 0.84 *** | 0.19 * | 0.08 | −0.13 |
| Li | 0.41 *** | 0.33 *** | 0.43 *** | 0.30 ** | 0.30 ** | 1 | 0.38 *** | 0.26 ** | 0.29 ** | 0.17 | 0.09 | −0.13 | −0.14 |
| Mg | 0.87 *** | 0.66 *** | 0.47 *** | 0.90 *** | 0.89 *** | 0.38 *** | 1 | 0.88 *** | 0.50 *** | 0.83 *** | 0.20 * | −0.01 | −0.19 * |
| Mn | 0.77 *** | 0.64 *** | 0.46 *** | 0.87 *** | 0.87 *** | 0.26 ** | 0.91 *** | 1 | 0.50 *** | 0.83 *** | 0.19 * | −0.08 | −0.25 ** |
| Ni | 0.32 *** | 0.27 ** | 0.37 *** | 0.42 *** | 0.38 *** | 0.30 *** | 0.46 *** | 0.34 *** | 1 | 0.38 *** | 0.21 * | −0.02 | −0.28 ** |
| Zn | 0.75 *** | 0.65 *** | 0.32 *** | 0.85 *** | 0.86 *** | 0.14 | 0.87 *** | 0.85 *** | 0.31 *** | 1 | 0.23 * | 0.07 | −0.19* |
| GPC | 0.31 *** | 0.22 * | 0.12 | 0.43 *** | 0.48 *** | −0.08 | 0.40 *** | 0.47 *** | 0.19 * | 0.51 *** | 1 | −0.37 *** | −0.36 *** |
| GY | 0.03 | 0.04 | 0.02 | −0.01 | −0.07 | 0.13 | 0.05 | 0.02 | −0.03 | −0.07 | −0.36 *** | 1 | NA |
| GYpTKW | −0.11 | −0.10 | 0.07 | −0.19 * | 0.22 * | 0.06 | −0.14 | −0.15 | −0.13 | −0.24 ** | −0.44 *** | NA | 1 |
| Gene Annotation (Gene ID) | Trait in Our Study a | Chromosome | PVE (%) b | Traits Influenced Based on the Annotations | References for the Association of Annotations with Traits |
|---|---|---|---|---|---|
| 2-oxoglutarate (2OG) and Fe (II)-dependent oxygenase superfamily protein (TraesCS2A01G519900-TraesCS2A01G520000) | Cu (1) | 2A | 5.3 | Fe, Mg | [33,40] |
| AP2-like ethylene-responsive transcription factor (TraesCS2A01G514200) | Cu (1) | 2A | 3.1 | As | |
| ATP synthase gamma chain (TraesCS6B01G117700) | Ca (1) | 6B | 19.9 | Fe, Zn | [41] |
| Chaperone protein dnaJ (TraesCS4B01G187600) | Zn (1) | 4B | 14.1 | Cd | [32] |
| F-box family protein domain (TraesCS6D01G360300, TraesCS3B01G479800, TraesCS3B01G111900, TraesCS6B01G268400, TraesCS6D01G064500, TraesCS2D01G106500, TraesCS4D01G333100) | Co (1), Li (1), Mg (2), Mn (1), Zn (2) | 2D, 3B, 4D, 6B, 6D | 1.8–25.2 | Fe | [34] |
| GDSL esterase/lipase (TraesCS5A01G096300) | Li (1) | 5A | 4.4 | Fe, Zn, Mn | [42] |
| Kinase family protein (TraesCS1B01G375400) | Li (1) | 1B | 13.5 | Cd, Zn | [35,36] |
| Leucine rich receptor-like protein kinase (TraesCS2D01G466400, TraesCS6B01G384300-TraesCS6B01G384400, TraesCS4A01G490700, TraesCS3B01G192500) | Ca (1), Li (1), Mg, Zn | 2D, 3B, 4A, 6B | 1.8–12.6 | Fe | [35,36,42] |
| MYB transcription factor (TraesCS6B01G053100) | Ca (1) | 6B | 9.9 | Cd, Fe, Zn | [37] |
| Na-translocating NADH-quinone reductase subunit A (TraesCS1A01G432900) | Fe (1) | 1A | 11.2 | Fe | [38] |
| No apical meristem (NAM) protein (TraesCS7A01G068200) | Ca (1) | 7A | 11.8 | Fe, Zn, N | [42] |
| Peroxidase (TraesCS6A01G081700) | Ca (1) | 6A | 9 | Cd | [43] |
| Phosphate translocator (TraesCS3B01G192400) | Zn (1) | 3B | 1.8 | P | [44] |
| Potassium transporter (TraesCS2D01G106600) | Mn (1) | 2D | 8.7 | K | [45] |
| Protein COBRA, putative (TraesCS4B01G187300) | Mn (1) | 4B | 13.4 | Al | [36] |
| Protein DETOXIFICATION (TraesCS3A01G300400) | Mg (1) | 3A | 14.6 | Fe | [46] |
| Protein ROOT HAIR DEFECTIVE 3 homolog (TraesCS1A01G003300-TraesCS1A01G003400) | Zn (1) | 1A | 3 | Cd | [32] |
| ROP guanine nucleotide exchange factor 10 (TraesCS4D01G333000) | Mg (1) | 4D | 7.9 | Fe | [36] |
| Universal stress protein family (TraesCS3B01G418000) | Ca (1) | 3B | 2.9 | Fe, Zn | [37] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhatta, M.; Baenziger, P.S.; Waters, B.M.; Poudel, R.; Belamkar, V.; Poland, J.; Morgounov, A. Genome-Wide Association Study Reveals Novel Genomic Regions Associated with 10 Grain Minerals in Synthetic Hexaploid Wheat. Int. J. Mol. Sci. 2018, 19, 3237. https://doi.org/10.3390/ijms19103237
Bhatta M, Baenziger PS, Waters BM, Poudel R, Belamkar V, Poland J, Morgounov A. Genome-Wide Association Study Reveals Novel Genomic Regions Associated with 10 Grain Minerals in Synthetic Hexaploid Wheat. International Journal of Molecular Sciences. 2018; 19(10):3237. https://doi.org/10.3390/ijms19103237
Chicago/Turabian StyleBhatta, Madhav, P. Stephen Baenziger, Brian M. Waters, Rachana Poudel, Vikas Belamkar, Jesse Poland, and Alexey Morgounov. 2018. "Genome-Wide Association Study Reveals Novel Genomic Regions Associated with 10 Grain Minerals in Synthetic Hexaploid Wheat" International Journal of Molecular Sciences 19, no. 10: 3237. https://doi.org/10.3390/ijms19103237
APA StyleBhatta, M., Baenziger, P. S., Waters, B. M., Poudel, R., Belamkar, V., Poland, J., & Morgounov, A. (2018). Genome-Wide Association Study Reveals Novel Genomic Regions Associated with 10 Grain Minerals in Synthetic Hexaploid Wheat. International Journal of Molecular Sciences, 19(10), 3237. https://doi.org/10.3390/ijms19103237