Assessment of Human Skin Gene Expression by Different Blends of Plant Extracts with Implications to Periorbital Skin Aging
Abstract
:1. Introduction
2. Results
2.1. BlendE: Modulation of Gene Expression (Antioxidants, Barrier Function, Lipid Metabolism/Transport)
2.2. BlendE: H2O2 Challenge, Oxidative Stress
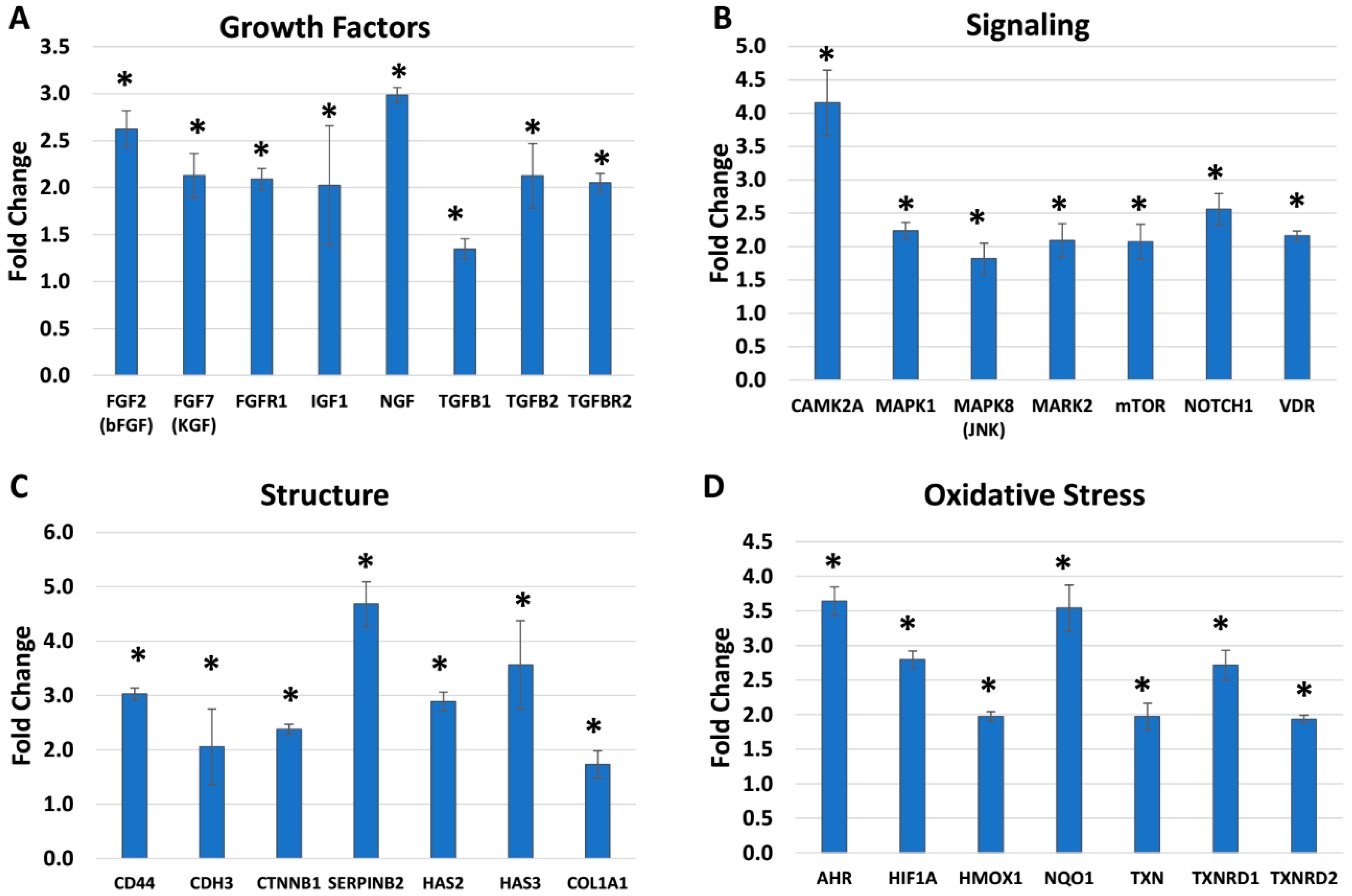
2.3. BlendIP: Skin Biomarker (Growth Factors, Signaling, Structural and Oxidative Stress)
2.4. BlendIP: Epigenetic Modulation
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. RNA Isolation and Real-Time Reverse Transcriptase Polymerase Chain Reaction
4.3. Data and Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABCA12 | ATP-binding cassette sub-family A member 12 |
| ACACB | acetyl-CoA carboxylase 2 |
| AHR | aryl hydrocarbon receptor |
| ANXA1 | annexin A1 |
| CAMPK2A | calcium/calmodulin dependent protein kinase II |
| CD | cell adhesion |
| CDH3 | cadherin |
| COL1A1 | collagen type 1 alpha 1 |
| CTNNB1 | beta-catenin |
| ECM | extracellular matrix |
| EFT | epidermal full-thickness |
| FABP | fatty acid binding protein |
| FBN | fibrillin |
| FGF | fibroblast growth factor |
| FLG | filaggrin |
| GPX1 | glutathione peroxidase 1 |
| H2O2 | hydrogen peroxide |
| HAS | hyaluronan synthase |
| HGF | hepatocyte growth factor |
| HIF1A | hypoxia-inducible factor 1 alpha |
| HMOX1 | heme oxygenase 1 |
| IGF | insulin growth factor |
| MAPK | mitogen-activated protein kinase |
| MARK2 | serine/threonine-protein kinase |
| MMP | matrix metalloprotease |
| mTOR | mechanistic target of rapamycin kinase |
| NGF | nerve growth factor |
| NOTCH1 | transmembrane protein 1 |
| NQO1 | NAD(P)H: quinone oxidoreductase |
| OCLN | occludin |
| ROS | radical oxygen species |
| SERPINB2 | plasminogen activator inhibitor type 2 |
| SOD | superoxide dismutase |
| ST14 | suppression of tumorigenicity 14 |
| TEWL | trans-epidermal water loss |
| TGF | transforming growth factor |
| TXN | thioredoxin |
| TXNRD1 | thioredoxin reductase 1 |
| VDR | vitamin D receptor |
| UGCG | UDP-glucose ceramide glucosyltransferase |
| UV | ultraviolet |
References
- Tobin, D.J. Introduction to skin aging. J. Tissue Viability 2017, 26, 37–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lephart, E.D. Equol’s anti-aging effects protects against environmental assaults by increasing Skin antioxidant defense and ECM proteins while decreasing oxidative stress and inflammation. Cosmetics 2018, 5, 16. [Google Scholar] [CrossRef]
- Kammeyer, A.; Luiten, R.M. Oxidation events and skin aging. Ageing Res. Rev. 2015, 21, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Firooz, A.; Rajabi-Estarabadi, A.; Zartab, H.; Pazhohi, N.; Fanian, F.; Janani, L. The influence of gender and age on the thickness and echo-density of skin. Skin Res. Technol. 2017, 23, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Rinnerthaler, M.; Bischof, J.; Streubel, M.K.; Trost, A.; Richter, K. Oxidative stress in aging human skin. Biomolecules 2015, 5, 545–589. [Google Scholar] [CrossRef] [PubMed]
- Bucay, V.D.; Doris, D. Adjunctive Skin Care of the Brow and Periorbital Region. Clin. Plast. Surg. 2013, 40, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Hillebrand, G.C. Facial wrinkling: The marquee clinical sing of aging skin. In Textbook of Skin Aging; Farage, M.A., Miller, H.I., Mailbach, H.I., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 911–918. [Google Scholar]
- Manaloto, R.M.; Alster, T.S. Periorbital rejuvenation: A review of dermatologic treatments. Dermatol. Surg. 1999, 25, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lambros, V. Observations on periorbital and midface aging. Plast. Reconstr. Surg. 2007, 120, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Baumann, L.S. Less-known botanical cosmeceuticals. Dermatol. Ther. 2007, 20, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Grunebaum, L.D.; Baumann, L.S. Nonprescription topical treatments for skin rejuvenation. Fac. Plast. Surg. 2014, 30, 3–11. [Google Scholar]
- Lephart, E.D. Skin aging and oxidative stress: Equol’s anti-aging effects via biochemical and molecular mechanisms. Ageing Res. Rev. 2016, 31, 36–54. [Google Scholar] [CrossRef] [PubMed]
- Lephart, E.D. Resveratrol, 4′ acetoxy resveratrol, R-equol, racemic equol. S-equol as cosmeceuticals to improve dermal health. Int. J. Mol. Sci. 2017, 18, 1193. [Google Scholar] [CrossRef] [PubMed]
- Panossian, A.; Wikman, G.; Sarris, J. Rosenroot (Rhodiola rosea): Traditional use, chemical composition, pharmacology and clinical efficacy. Phytomedicine 2010, 17, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Hancke, J.L.; Burgos, R.A.; Ahumada, F. Schisandra chinensis (Turcz.) Baill. Fitoterapia 1999, 70, 451–471. [Google Scholar] [CrossRef]
- Gronniger, E.; Weber, B.; Heil, O.; Peters, N.; Stab, F.; Wenck, H.; Korn, B.; Winnefeld, M.; Lyko, F. Aging and chronic sun exposure cause distinct epigenetic changes in human skin. PLoS Genet. 2010, 6, e1000971. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.R.; Nam, K.M.; Lee, H.S.; Yang, S.H.; Kim, Y.S.; Lee, J.; Date, A.; Toyama, K.; Park, K.C. Phlorizin, an Active Ingredient of Eleutherococcus senticosus, Increases Proliferative Potential of Keratinocytes with Inhibition of MiR135b and Increased Expression of Type IV Collagen. Oxid. Med. Cell. Longev. 2016, 2016, 3859721. [Google Scholar] [CrossRef] [PubMed]
- Penta, D.; Somashekar, B.S.; Meeran, S.M. Epigenetics of skin cancer: Interventions by selected bioactive phytochemicals. Photodermatol. Photoimmunol. Photomed. 2018, 34, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.Y.; Li, L.; Wang, M.J.; Chen, X.M.; Huang, Q.C.; Lu, C.J. An exploration of the role of microRNAs in psoriasis a systemic review of the literature. Medicine 2015, 94, e2030. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Zhang, W. MicroRNAs in normal and psoriastic skin. Physiol. Gemom. 2014, 46, 122. [Google Scholar]
- Dorni, A.I.C.; Amalraj, A.; Gopi, S.; Varma, K.; Anjana, S.N. Novel cosmeceuticals from plants—An industry guided review. J. Appl. Res. Med. Aromat. Plants 2017, 7, 1–26. [Google Scholar]
- Lupa, D.M.; Fakfalah, F.; Safferling, K.; Boukamp, P.; Poschmann, G.; Volpo, E.; Gotz-Rosch, C.; Bernerd, F.; Haag, L.; Huebenthal, U.; et al. Characterization of skin aging—Associated secreted proteins (SSAAP) produced by dermal fibroblasts isolated from intrinsically aged human skin. J. Investig. Dermatol. 2015, 135, 1954–1968. [Google Scholar] [CrossRef] [PubMed]
- The Human Protein Atlas: The Human Tissues. The Skin-Specific Proteome. Available online: https://www.proteinsatlas.org/humanproteone/skin (accessed on 15 October 2018).
- Shindo, Y.; Witt, E.; Han, D.; Epstein, W.; Packer, L. Enzymic and non-enzymic antioxidants in epidermis and dermis in human skin. J. Investig. Dermatol. 1994, 102, 122–124. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Kong, Y.; Zhang, H. Oxidative stress, mitochondrial dysfunction, and aging. J. Signal Transduct. 2012, 2012, 646354. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.J.; Kim, B.; Lee, Y.S.; Kim, T.Y. Role of Superoxide Dismutase 3 in Skin Inflammation. J. Dermatol. Sci. 2012, 67, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Gopaul, R.; Knaggs, H.; Lephart, E.D. Biochemical investigation and gene analysis of equol: A plant and soy-derived isoflavonoid with anti-aging and antioxidant properties with potential human skin applications. Biofactors 2012, 38, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.R. The glutathione peroxidases. Cell. Mol. Life Sci. 2001, 57, 1825–1835. [Google Scholar] [CrossRef]
- Furue, M.; Takahara, M.; Nakahara, T.; Uchi, H. Role of Ahr/ARNT system in skin homeostasis. Arch. Dermatol. Res. 2014, 306, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Domingo, D.S.; Camouse, M.M.; Hsia, A.H.; Matsui, M.; Maes, D.; Ward, N.L.; Cooper, K.D.; Baron, E.D. Anti-angiogenic effects of epigallocatechin-3-gallate in human skin. Int. J. Clin. Exp. Pathol. 2010, 3, 705–709. [Google Scholar] [PubMed]
- Tu, Y.; Quan, T. Oxidative Stress and Human Skin Connective Tissue Aging. Cosmetics 2016, 3, 28. [Google Scholar] [CrossRef]
- Akiyama, M. Corneocyte lipid envelope (CLE), the key structure for skin barrier function and ichthyosis pathogenesis. J. Dermatol. Sci. 2017, 88, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Elias, P.M. Skin barrier function. Curr. Allergy Asthma Rep. 2008, 8, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, M. The roles of ABCA12 in epidermal lipid barrier formation and keratinocyte differentiation. Biochim. Biophys. Acta 2014, 1841, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Buraczewska, I.; Berne, B.; Lindberg, M.; Loden, M.; Torma, H. Moisturizers change the mRNA expression of enzymes synthesizing skin barrier lipids. Arch. Dermatol. Res. 2009, 301, 587. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Khnykin, D. Fatty acid transporters in skin development, function and disease. Biochim. Biophys. Acta 2014, 1841, 362. [Google Scholar] [CrossRef] [PubMed]
- Fraga, M.F.; Esteller, M. Epigenetics and aging: The targets and the marks. Trends Genet. 2007, 23, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Mancini, M.; Lena, A.M.; Saintigny, G.; Mahe, C.; Di Daniele, N.; Melino, G.; Candi, E. MicroRNAs in human skin aging. Ageing Res. Rev. 2014, 17, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, P.; Ceccoli, J. Advances in the applications and impact of microRNAs as therapies for skin diseases. Biodrugs 2017, 31, 423–438. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, S. Epigenetic alterations in aging. J. Appl. Physiol. 2010, 109, 586–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Botchkareva, N.V. MicroRNA/mRNA regulatory networks in the control of skin development and regeneration. Cell Cycle 2012, 11, 468–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syed, D.N.; Khan, M.I.; Shabbir, M.; Mukhtar, H. MicroRNAs in skin response to UV radiation. Curr. Drug Targets 2013, 14, 1128–1134. [Google Scholar] [CrossRef] [PubMed]
- Meisgen, F.; Xu Landen, N.; Wang, A.; Rethi, B.; Bouez, C.; Zuccolo, M.; Gueniche, A.; Stahle, M.; Sonkoly, E.; Breton, L.; et al. MiR-146a negatively regulates TLR2-induced inflammatory responses in keratinocytes. J. Investig. Dermatol. 2014, 134, 1931–1940. [Google Scholar] [CrossRef] [PubMed]
- Cha, H.J.; Lee, K.S.; Lee, G.T.; Lee, K.K.; Hong, J.T.; Lee, S.N.; Jang, H.H.; Lee, J.H.; Park, I.H.; Kim, Y.R.; et al. Altered miRNA expression profiles are involved in the protective effects of troxerutin against ultraviolet B radiation in normal human dermal fibroblasts. Int. J. Mol. Med. 2014, 33, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Kappil, M.; Chen, J. Environmental exposures in utero and microRNA. Curr. Opin. Pediatr. 2014, 26, 243–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naidoo, K.; Birch-Machin, M.A. Oxidative stress and ageing: The influence of environmental pollution, sunlight and diet on skin. Cosmetics 2017, 4, 4. [Google Scholar] [CrossRef]
- Hamer, M.A.; Pardo, L.M.; Jacobs, L.C.; Ikram, M.A.; Laven, J.S.; Kayser, M.; Hollestein, L.M.; Gunn, D.A.; Nijsten, T. Lifestyle and physiological factors associated with facial wrinkling in men and women. J. Investig. Dermatol. 2017, 137, 1692–1699. [Google Scholar] [CrossRef] [PubMed]
- Analysis of Variance from Summary Data. Available online: http://statpages.info/anova1sm.html (accessed on 11 September 2018).



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Namkoong, J.; Kern, D.; Knaggs, H.E. Assessment of Human Skin Gene Expression by Different Blends of Plant Extracts with Implications to Periorbital Skin Aging. Int. J. Mol. Sci. 2018, 19, 3349. https://doi.org/10.3390/ijms19113349
Namkoong J, Kern D, Knaggs HE. Assessment of Human Skin Gene Expression by Different Blends of Plant Extracts with Implications to Periorbital Skin Aging. International Journal of Molecular Sciences. 2018; 19(11):3349. https://doi.org/10.3390/ijms19113349
Chicago/Turabian StyleNamkoong, Jin, Dale Kern, and Helen E. Knaggs. 2018. "Assessment of Human Skin Gene Expression by Different Blends of Plant Extracts with Implications to Periorbital Skin Aging" International Journal of Molecular Sciences 19, no. 11: 3349. https://doi.org/10.3390/ijms19113349



