A Two-Step Approach for the Design and Generation of Nanobodies
Abstract
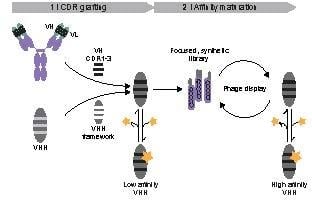
:1. Introduction
2. Results
2.1. Design of Toxin- and Hapten-Specific VHH Grafts
2.2. Evaluation of Antigen Binding
2.3. Design of a Synthetic VHH Library
2.4. Selection of Fluorescein-Binding VHH from the Synthetic Library
2.5. Characterization of a Selected Fluorescein-Binding VHH
3. Discussion
4. Materials and Methods
4.1. Graft Construction
4.2. Protein Production and Purification
4.3. Synthesis of Flu-BSA
4.4. Analytical Methods
4.5. Synthetic Flu-VHH Library and Phage Display
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BoNT/A-LC | Botulinum neurotoxin serotype A light chain |
| BSA | Bovine serum albumin |
| CDR | Complementarity-determining region |
| ELISA | Enzyme-linked immunosorbent assay |
| EMA | European Medicines Agency |
| Fab | Fragment antigen-binding |
| FDA | Food and Drug Administration |
| Flu | Fluorescein |
| HCAb | Heavy-chain-only antibody |
| sdAb | Single-domain antibody |
| scFv | single-chain variable fragment |
| VH | Heavy chain variable domain |
| VHH | variable domain of camelid heavy-chain-only antibodies |
| VL | Light chain variable domain |
| VNAR | Variable domain of the immunoglobulin new antigen receptor (IgNAR) from sharks |
References
- Chatenoud, L.; Bluestone, J.A. CD3-specific antibodies: A portal to the treatment of autoimmunity. Nat. Rev. Immunol. 2007, 7, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Sliwkowski, M.X.; Mellman, I. Antibody Therapeutics in Cancer. Science 2013, 341, 1192–1198. [Google Scholar] [CrossRef] [PubMed]
- Marasco, W.A.; Sui, J. The growth and potential of human antiviral monoclonal antibody therapeutics. Nat. Biotechnol. 2007, 25, 1421–1434. [Google Scholar] [CrossRef] [PubMed]
- Kaplon, H.; Reichert, J.M. Antibodies to watch in 2018. MAbs 2018, 10, 183–203. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.N.; Wormald, M.R.; Sim, R.B.; Rudd, P.M.; Dwek, R.A. The Impact of Glycosylation on the Biological Function and Structure of Human Immunoglobulins. Annu. Rev. Immunol. 2007, 25, 21–50. [Google Scholar] [CrossRef] [PubMed]
- Nord, K.; Gunneriusson, E.; Ringdahl, J.; Ståhl, S.; Uhlén, M.; Nygren, P.-Å. Binding proteins selected from combinatorial libraries of an α-helical bacterial receptor domain. Nat. Biotechnol. 1997, 15, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Ståhl, S.; Gräslund, T.; Eriksson Karlström, A.; Frejd, F.Y.; Nygren, P.-Å.; Löfblom, J. Affibody Molecules in Biotechnological and Medical Applications. Trends Biotechnol. 2017, 35, 691–712. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, J.; Hantschel, O.; Grebien, F.; Kaupe, I.; Bennett, K.L.; Barkinge, J.; Jones, R.B.; Koide, A.; Superti-Furga, G.; Koide, S. A potent and highly specific FN3 monobody inhibitor of the Abl SH2 domain. Nat. Struct. Mol. Biol. 2010, 17, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Beste, G.; Schmidt, F.S.; Stibora, T.; Skerra, A. Small antibody-like proteins with prescribed ligand specificities derived from the lipocalin fold. Proc. Natl. Acad. Sci. USA 1999, 96, 1898–1903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Yang, Y.; Dikici, E.; Deo, S.K.; Daunert, S. Beyond Antibodies as Binding Partners: The Role of Antibody Mimetics in Bioanalysis. Annu. Rev. Anal. Chem. 2017, 10, 293–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dmitriev, O.Y.; Lutsenko, S.; Muyldermans, S. Nanobodies as Probes for Protein Dynamics in Vitro and in Cells. J. Biol. Chem. 2016, 291, 3767–3775. [Google Scholar] [CrossRef] [PubMed]
- Bever, C.S.; Dong, J.-X.; Vasylieva, N.; Barnych, B.; Cui, Y.; Xu, Z.-L.; Hammock, B.D.; Gee, S.J. VHH antibodies: Emerging reagents for the analysis of environmental chemicals. Anal. Bioanal. Chem. 2016, 408, 5985–6002. [Google Scholar] [CrossRef] [PubMed]
- Muyldermans, S.; Baral, T.N.; Retamozzo, V.C.; De Baetselier, P.; De Genst, E.; Kinne, J.; Leonhardt, H.; Magez, S.; Nguyen, V.K.; Revets, H.; et al. Camelid immunoglobulins and nanobody technology. Vet. Immunol. Immunopathol. 2009, 128, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Muyldermans, S. Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [PubMed]
- Ingram, J.R.; Schmidt, F.I.; Ploegh, H.L. Exploiting Nanobodies’ Singular Traits. Annu. Rev. Immunol. 2018, 36, 695–715. [Google Scholar] [CrossRef] [PubMed]
- Broisat, A.; Hernot, S.; Toczek, J.; De Vos, J.; Riou, L.M.; Martin, S.; Ahmadi, M.; Thielens, N.; Wernery, U.; Caveliers, V.; et al. Nanobodies Targeting Mouse/Human VCAM1 for the Nuclear Imaging of Atherosclerotic Lesions. Circ. Res. 2012, 110, 927–937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xavier, C.; Blykers, A.; Vaneycken, I.; D’Huyvetter, M.; Heemskerk, J.; Lahoutte, T.; Devoogdt, N.; Caveliers, V. 18F-nanobody for PET imaging of HER2 overexpressing tumors. Nucl. Med. Biol. 2016, 43, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Gulati, S.; Jin, H.; Masuho, I.; Orban, T.; Cai, Y.; Pardon, E.; Martemyanov, K.A.; Kiser, P.D.; Stewart, P.L.; Ford, C.P.; et al. Targeting G protein-coupled receptor signaling at the G protein level with a selective nanobody inhibitor. Nat. Commun. 2018, 9, 1996. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Duarte, J.N.; Ling, J.; Li, Z.; Guzman, J.S.; Ploegh, H.L. Structurally Defined αMHC-II Nanobody-Drug Conjugates: A Therapeutic and Imaging System for B-Cell Lymphoma. Angew. Chem. Int. Ed. 2016, 55, 2416–2420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Driel, P.B.A.A.; Boonstra, M.C.; Slooter, M.D.; Heukers, R.; Stammes, M.A.; Snoeks, T.J.A.; de Bruijn, H.S.; van Diest, P.J.; Vahrmeijer, A.L.; van Bergen en Henegouwen, P.M.P.; et al. EGFR targeted nanobody-photosensitizer conjugates for photodynamic therapy in a pre-clinical model of head and neck cancer. J. Control. Release 2016, 229, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Peyvandi, F.; Scully, M.; Kremer Hovinga, J.A.; Knöbl, P.; Cataland, S.; De Beuf, K.; Callewaert, F.; De Winter, H.; Zeldin, R.K. Caplacizumab reduces the frequency of major thromboembolic events, exacerbations and death in patients with acquired thrombotic thrombocytopenic purpura. J. Thromb. Haemost. 2017, 15, 1448–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peyvandi, F.; Scully, M.; Kremer Hovinga, J.A.; Cataland, S.; Knöbl, P.; Wu, H.; Artoni, A.; Westwood, J.-P.; Mansouri Taleghani, M.; Jilma, B.; et al. Caplacizumab for Acquired Thrombotic Thrombocytopenic Purpura. N. Engl. J. Med. 2016, 374, 511–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco, E.J.; Sonneson, G.J.; DeLegge, T.J.; Hofstetter, H.; Horn, J.R.; Hofstetter, O. Production and characterization of a genetically engineered anti-caffeine camelid antibody and its use in immunoaffinity chromatography. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2010, 878, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Duc, T.; Peeters, E.; Muyldermans, S.; Charlier, D.; Hassanzadeh-Ghassabeh, G. Nanobody®-based chromatin immunoprecipitation/micro-array analysis for genome-wide identification of transcription factor DNA binding sites. Nucleic Acids Res. 2013, 41, e59. [Google Scholar] [CrossRef] [PubMed]
- Pardon, E.; Laeremans, T.; Triest, S.; Rasmussen, S.G.F.; Wohlkönig, A.; Ruf, A.; Muyldermans, S.; Hol, W.G.J.; Kobilka, B.K.; Steyaert, J. A general protocol for the generation of Nanobodies for structural biology. Nat. Protoc. 2014, 9, 674–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Song, H.; Chen, Q.; Yu, J.; Xian, M.; Nian, R.; Feng, D. Recent advances in the selection and identification of antigen-specific nanobodies. Mol. Immunol. 2018, 96, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Maass, D.R.; Sepulveda, J.; Pernthaner, A.; Shoemaker, C.B. Alpaca (Lama pacos) as a convenient source of recombinant camelid heavy chain antibodies (VHHs). J. Immunol. Methods 2007, 324, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Rothbauer, U.; Zolghadr, K.; Tillib, S.; Nowak, D.; Schermelleh, L.; Gahl, A.; Backmann, N.; Conrath, K.; Muyldermans, S.; Cardoso, M.C.; et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 2006, 3, 887–889. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhu, M.; Ma, L.; Yan, J.; Lu, X.; Shen, Y.; Wan, Y. Generation of Small Single Domain Nanobody Binders for Sensitive Detection of Testosterone by Electrochemical Impedance Spectroscopy. ACS Appl. Mater. Interfaces 2016, 8, 13830–13839. [Google Scholar] [CrossRef] [PubMed]
- Sheedy, C.; Yau, K.Y.F.; Hirama, T.; MacKenzie, C.R.; Hall, J.C. Selection, characterization, and CDR shuffling of naive llama single-domain antibodies selected against auxin and their cross-reactivity with auxinic herbicides from four chemical families. J. Agric. Food Chem. 2006, 54, 3668–3678. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Rueda, N.; Behar, G.; Ferré, V.; Pugnière, M.; Roquet, F.; Gastinel, L.; Jacquot, C.; Aubry, J.; Baty, D.; Barbet, J.; Birklé, S. Generation of llama single-domain antibodies against methotrexate, a prototypical hapten. Mol. Immunol. 2007, 44, 1680–1690. [Google Scholar] [CrossRef] [PubMed]
- Bakherad, H.; Mousavi Gargari, S.L.; Rasooli, I.; RajabiBazl, M.; Mohammadi, M.; Ebrahimizadeh, W.; Safaee Ardakani, L.; Zare, H. In Vivo Neutralization of Botulinum Neurotoxins Serotype E with Heavy-chain Camelid Antibodies (VHH). Mol. Biotechnol. 2013, 55, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Hmila, I.; Abdallah R, B.A.-B.; Saerens, D.; Benlasfar, Z.; Conrath, K.; El Ayeb, M.; Muyldermans, S.; Bouhaouala-Zahar, B. VHH, bivalent domains and chimeric Heavy chain-only antibodies with high neutralizing efficacy for scorpion toxin AahI’. Mol. Immunol. 2008, 45, 3847–3856. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, J.M.; Mukherjee, J.; Leysath, C.E.; Debatis, M.; Ofori, K.; Baldwin, K.; Boucher, C.; Peters, R.; Beamer, G.; Sheoran, A.; et al. A Single VHH-Based Toxin-Neutralizing Agent and an Effector Antibody Protect Mice against Challenge with Shiga Toxins 1 and 2. Infect. Immun. 2013, 81, 4592–4603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tremblay, J.M.; Kuo, C.-L.; Abeijon, C.; Sepulveda, J.; Oyler, G.; Hu, X.; Jin, M.M.; Shoemaker, C.B. Camelid single domain antibodies (VHHs) as neuronal cell intrabody binding agents and inhibitors of Clostridium botulinum neurotoxin (BoNT) proteases. Toxicon 2010, 56, 990–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabares-da Rosa, S.; Rossotti, M.; Carleiza, C.; Carrión, F.; Pritsch, O.; Ahn, K.C.; Last, J.A.; Hammock, B.D.; González-Sapienza, G. Competitive Selection from Single Domain Antibody Libraries Allows Isolation of High-Affinity Antihapten Antibodies That Are Not Favored in the llama Immune Response. Anal. Chem. 2011, 83, 7213–7220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; McCoy, M.R.; Majkova, Z.; Dechant, J.E.; Gee, S.J.; Tabares-da Rosa, S.; González-Sapienza, G.G.; Hammock, B.D. Isolation of Alpaca Anti-Hapten Heavy Chain Single Domain Antibodies for Development of Sensitive Immunoassay. Anal. Chem. 2012, 84, 1165–1171. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Li, G.; Hu, Y.; Ou, W.; Wan, Y. Construction of a synthetic phage-displayed Nanobody library with CDR3 regions randomized by trinucleotide cassettes for diagnostic applications. J. Transl. Med. 2014, 12, 343. [Google Scholar] [CrossRef] [PubMed]
- Moutel, S.; Bery, N.; Bernard, V.; Keller, L.; Lemesre, E.; De Marco, A.; Ligat, L.; Rain, J.C.; Favre, G.; Olichon, A.; et al. NaLi-H1: A universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. Elife 2016. [Google Scholar] [CrossRef] [PubMed]
- Yau, K.Y.F.; Groves, M.A.T.; Li, S.; Sheedy, C.; Lee, H.; Tanha, J.; MacKenzie, C.R.; Jermutus, L.; Hall, J.C. Selection of hapten-specific single-domain antibodies from a non-immunized llama ribosome display library. J. Immunol. Methods 2003, 281, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-F.; Ho, M. Humanization of rabbit monoclonal antibodies via grafting combined Kabat/IMGT/Paratome complementarity-determining regions: Rationale and examples. MAbs 2017, 9, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Carter, P.; Presta, L.; Gorman, C.M.; Ridgway, J.B.; Henner, D.; Wong, W.L.; Rowland, A.M.; Kotts, C.; Carver, M.E.; Shepard, H.M. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc. Natl. Acad. Sci. USA 1992, 89, 4285–4289. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.G.; Matthews, D.J.; Jones, T. Humanising Antibodies by CDR Grafting. In Antibody Engineering; Kontermann, R., Dübel, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 319–339. ISBN 978-3-642-01144-3. [Google Scholar]
- Saerens, D.; Pellis, M.; Loris, R.; Pardon, E.; Dumoulin, M.; Matagne, A.; Wyns, L.; Muyldermans, S.; Conrath, K. Identification of a universal VHH framework to graft non-canonical antigen-binding loops of camel single-domain antibodies. J. Mol. Biol. 2005, 352, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Vincke, C.; Loris, R.; Saerens, D.; Martinez-Rodriguez, S.; Muyldermans, S.; Conrath, K. General Strategy to Humanize a Camelid Single-domain Antibody and Identification of a Universal Humanized Nanobody Scaffold. J. Biol. Chem. 2009, 284, 3273–3284. [Google Scholar] [CrossRef] [PubMed]
- Fanning, S.W.; Horn, J.R. An anti-hapten camelid antibody reveals a cryptic binding site with significant energetic contributions from a nonhypervariable loop. Protein Sci. 2011, 20, 1196–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ewert, S.; Honegger, A.; Plückthun, A. Stability improvement of antibodies for extracellular and intracellular applications: CDR grafting to stable frameworks and structure-based framework engineering. Methods 2004, 34, 184–199. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, J.; Mukherjee, J.; Tzipori, S.; Simpson, L.L.; Shoemaker, C.B. Efficient Serum Clearance of Botulinum Neurotoxin Achieved Using a Pool of Small Antitoxin Binding Agents. Infect. Immun. 2010, 78, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, T.J.; Williams, A.J.; Pritchard, K.; Osbourn, J.K.; Pope, A.R.; Earnshaw, J.C.; McCafferty, J.; Hodits, R.A.; Wilton, J.; Johnson, K.S. Human Antibodies with Sub-nanomolar Affinities Isolated from a Large Non-immunized Phage Display Library. Nat. Biotechnol. 1996, 14, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Kirchhofer, A.; Helma, J.; Schmidthals, K.; Frauer, C.; Cui, S.; Karcher, A.; Pellis, M.; Muyldermans, S.; Casas-Delucchi, C.S.; Cardoso, M.C.; et al. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 2010, 17, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Kabat, E.A.; Wu, T.T.; Foeller, C.; Perry, H.M.; Gottesman, K.S. Sequences of Proteins of Immunological Interest, 5th ed.; United States Department of Health and Human Services, Public Health Service, and NIH: Bethesda, MD, USA, 1991.
- Foote, J.; Winter, G. Antibody framework residues affecting the conformation of the hypervariable loops. J. Mol. Biol. 1992, 224, 487–499. [Google Scholar] [CrossRef]
- Crooks, G.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honegger, A.; Plückthun, A. Yet Another Numbering Scheme for Immunoglobulin Variable Domains: An Automatic Modeling and Analysis Tool. J. Mol. Biol. 2001, 309, 657–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fellouse, F.A.; Li, B.; Compaan, D.M.; Peden, A.A.; Hymowitz, S.G.; Sidhu, S.S. Molecular Recognition by a Binary Code. J. Mol. Biol. 2005, 348, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Marks, J.D.; Griffiths, A.D.; Malmqvist, M.; Clackson, T.P.; Bye, J.M.; Winter, G. By-Passing Immunization: Building High Affinity Human Antibodies by Chain Shuffling. Nat. Biotechnol. 1992, 10, 779–783. [Google Scholar] [CrossRef]
- Tu, Z.; Xu, Y.; He, Q.; Fu, J.; Liu, X.; Tao, Y. Isolation and characterisation of deoxynivalenol affinity binders from a phage display library based on single-domain camelid heavy chain antibodies (VHHs). Food Agric. Immunol. 2012, 23, 123–131. [Google Scholar] [CrossRef]
- Qiu, Y.-L.; He, Q.-H.; Xu, Y.; Bhunia, A.K.; Tu, Z.; Chen, B.; Liu, Y.-Y. Deoxynivalenol-mimic nanobody isolated from a naïve phage display nanobody library and its application in immunoassay. Anal. Chim. Acta 2015, 887, 201–208. [Google Scholar] [CrossRef] [PubMed]
- de Haard, H.J.; van Neer, N.; Reurs, A.; Hufton, S.E.; Roovers, R.C.; Henderikx, P.; de Bruïne, A.P.; Arends, J.; Hoogenboom, H.R. A Large Non-immunized Human Fab Fragment Phage Library That Permits Rapid Isolation and Kinetic Analysis of High Affinity Antibodies. J. Biol. Chem. 1999, 274, 18218–18230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.V.; Liang, W.-C.; Dennis, M.S.; Eigenbrot, C.; Sidhu, S.S.; Fuh, G. High-affinity Human Antibodies from Phage-displayed Synthetic Fab Libraries with a Single Framework Scaffold. J. Mol. Biol. 2004, 340, 1073–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fellouse, F.A.; Esaki, K.; Birtalan, S.; Raptis, D.; Cancasci, V.J.; Koide, A.; Jhurani, P.; Vasser, M.; Wiesmann, C.; Kossiakoff, A.A.; et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J. Mol. Biol. 2007, 373, 924–940. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, S.S.; Li, B.; Chen, Y.; Fellouse, F.A.; Eigenbrot, C.; Fuh, G. Phage-displayed Antibody Libraries of Synthetic Heavy Chain Complementarity Determining Regions. J. Mol. Biol. 2004, 338, 299–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knappik, A.; Ge, L.; Honegger, A.; Pack, P.; Fischer, M.; Wellnhofer, G.; Hoess, A.; Wölle, J.; Plückthun, A.; Virnekäs, B. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides 1 1Edited by I.A. Wilson. J. Mol. Biol. 2000, 296, 57–86. [Google Scholar] [CrossRef] [PubMed]
- Philibert, P.; Stoessel, A.; Wang, W.; Sibler, A.-P.; Bec, N.; Larroque, C.; Saven, J.G.; Courtête, J.; Weiss, E.; Martineau, P. A focused antibody library for selecting scFvs expressed at high levels in the cytoplasm. BMC Biotechnol. 2007, 7, 81. [Google Scholar] [CrossRef] [PubMed]
- McMahon, C.; Baier, A.S.; Pascolutti, R.; Wegrecki, M.; Zheng, S.; Ong, J.X.; Erlandson, S.C.; Hilger, D.; Rasmussen, S.G.F.; Ring, A.M.; et al. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat. Struct. Mol. Biol. 2018, 25, 289–296. [Google Scholar] [CrossRef] [PubMed]
- MacCallum, R.M.; Martin, A.C.R.; Thornton, J.M. Antibody-antigen Interactions: Contact Analysis and Binding Site Topography. J. Mol. Biol. 1996, 262, 732–745. [Google Scholar] [CrossRef] [PubMed]
- Persson, H.; Lantto, J.; Ohlin, M. A Focused Antibody Library for Improved Hapten Recognition. J. Mol. Biol. 2006, 357, 607–620. [Google Scholar] [CrossRef] [PubMed]







© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wagner, H.J.; Wehrle, S.; Weiss, E.; Cavallari, M.; Weber, W. A Two-Step Approach for the Design and Generation of Nanobodies. Int. J. Mol. Sci. 2018, 19, 3444. https://doi.org/10.3390/ijms19113444
Wagner HJ, Wehrle S, Weiss E, Cavallari M, Weber W. A Two-Step Approach for the Design and Generation of Nanobodies. International Journal of Molecular Sciences. 2018; 19(11):3444. https://doi.org/10.3390/ijms19113444
Chicago/Turabian StyleWagner, Hanna J., Sarah Wehrle, Etienne Weiss, Marco Cavallari, and Wilfried Weber. 2018. "A Two-Step Approach for the Design and Generation of Nanobodies" International Journal of Molecular Sciences 19, no. 11: 3444. https://doi.org/10.3390/ijms19113444