Identification of Potential Key Genes Associated with Adipogenesis through Integrated Analysis of Five Mouse Transcriptome Datasets
Abstract
:1. Introduction
2. Results
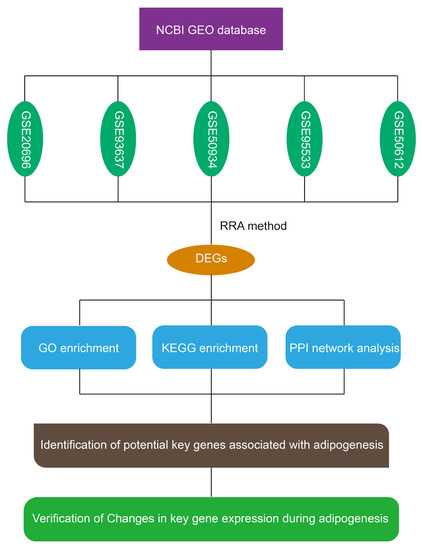
2.1. Expression Profile Datasets and Identification of Differentially Expressed Genes (DEGs) Associated with Adipogenesis
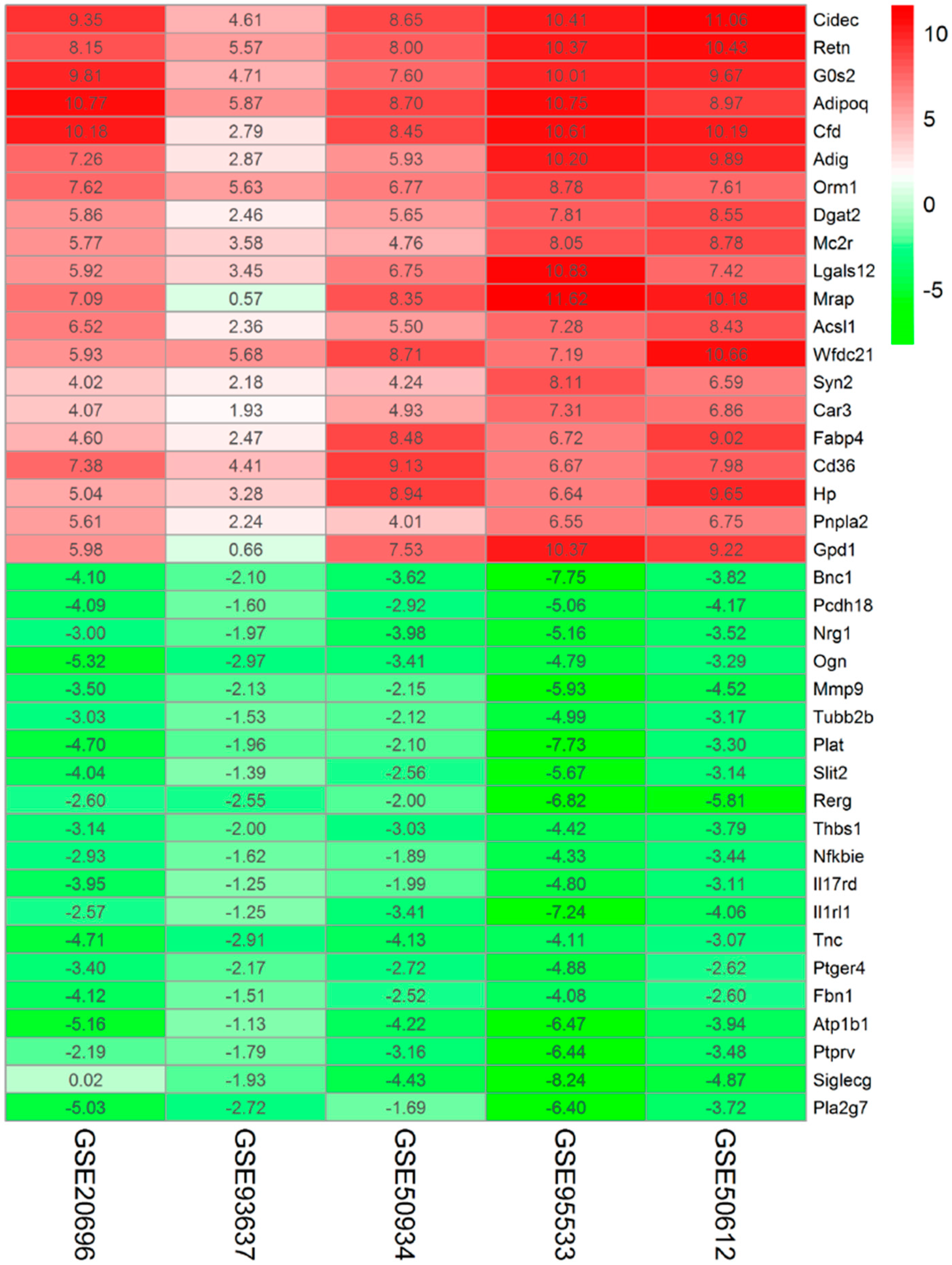
2.2. Identification of DEGs Associated with Adipogenesis Using Integrated Bioinformatics
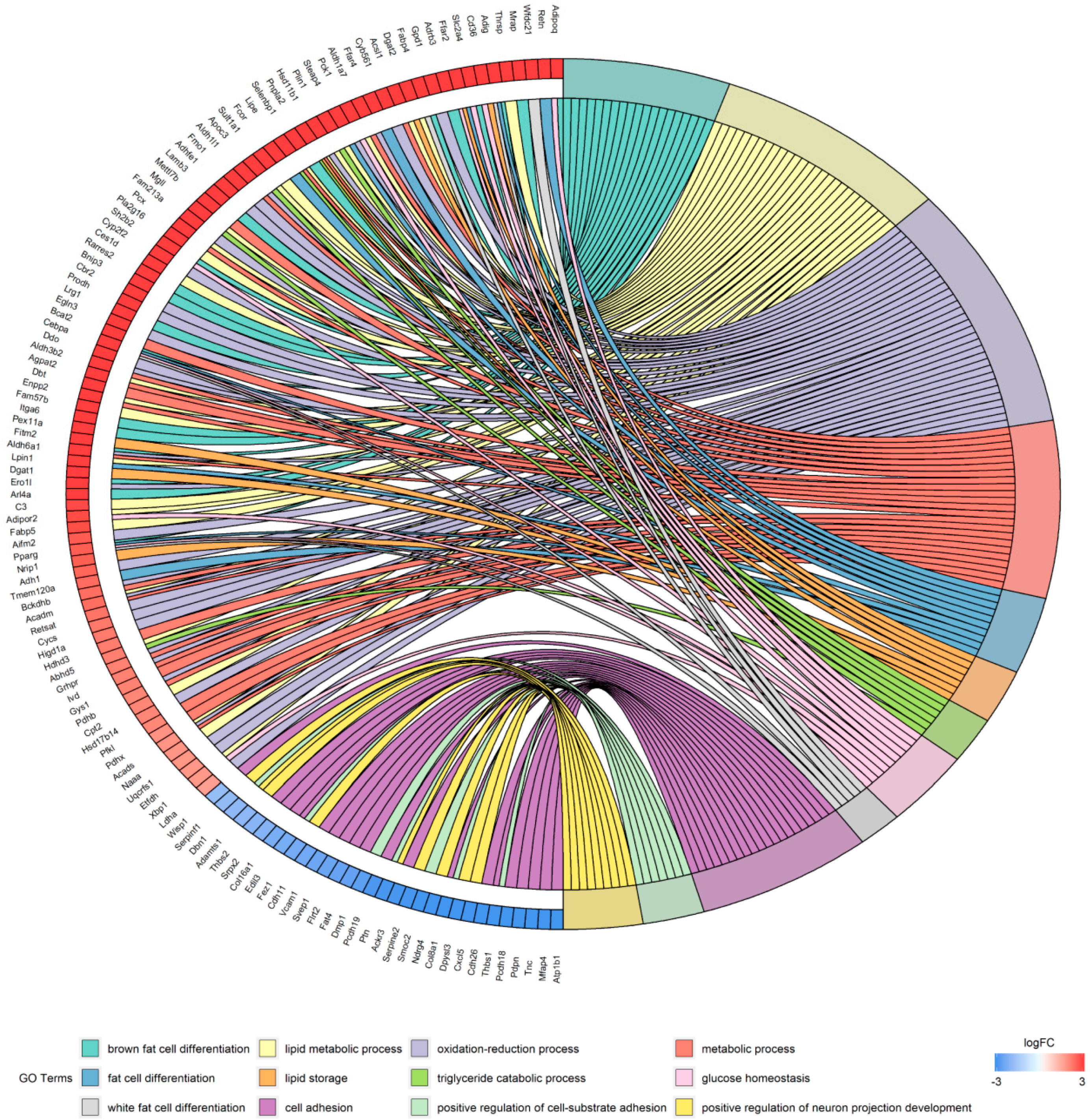
2.3. Enrichment Analysis of the Gene Ontology (GO) Terms of the DEGs
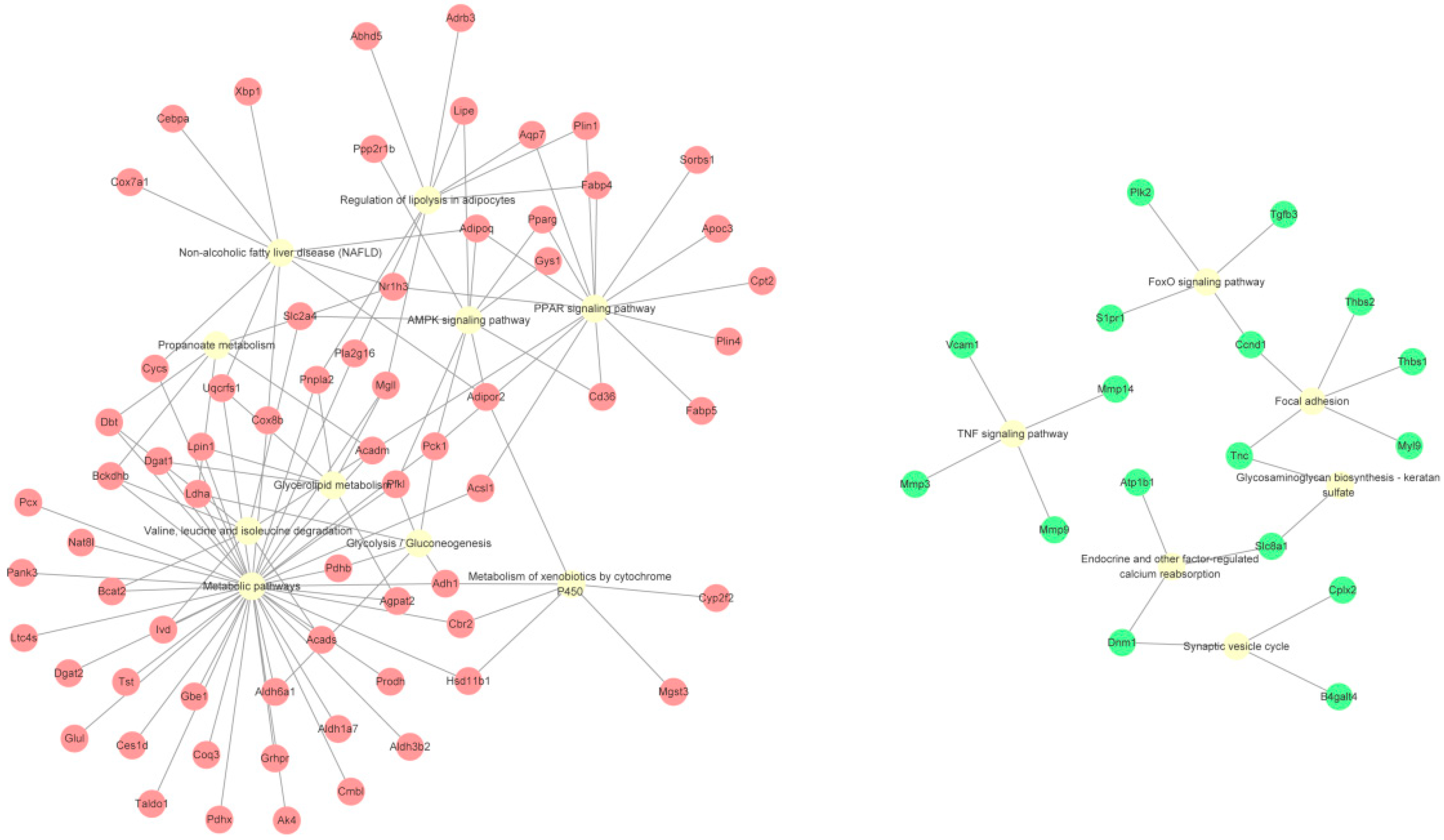
2.4. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis of DEGs
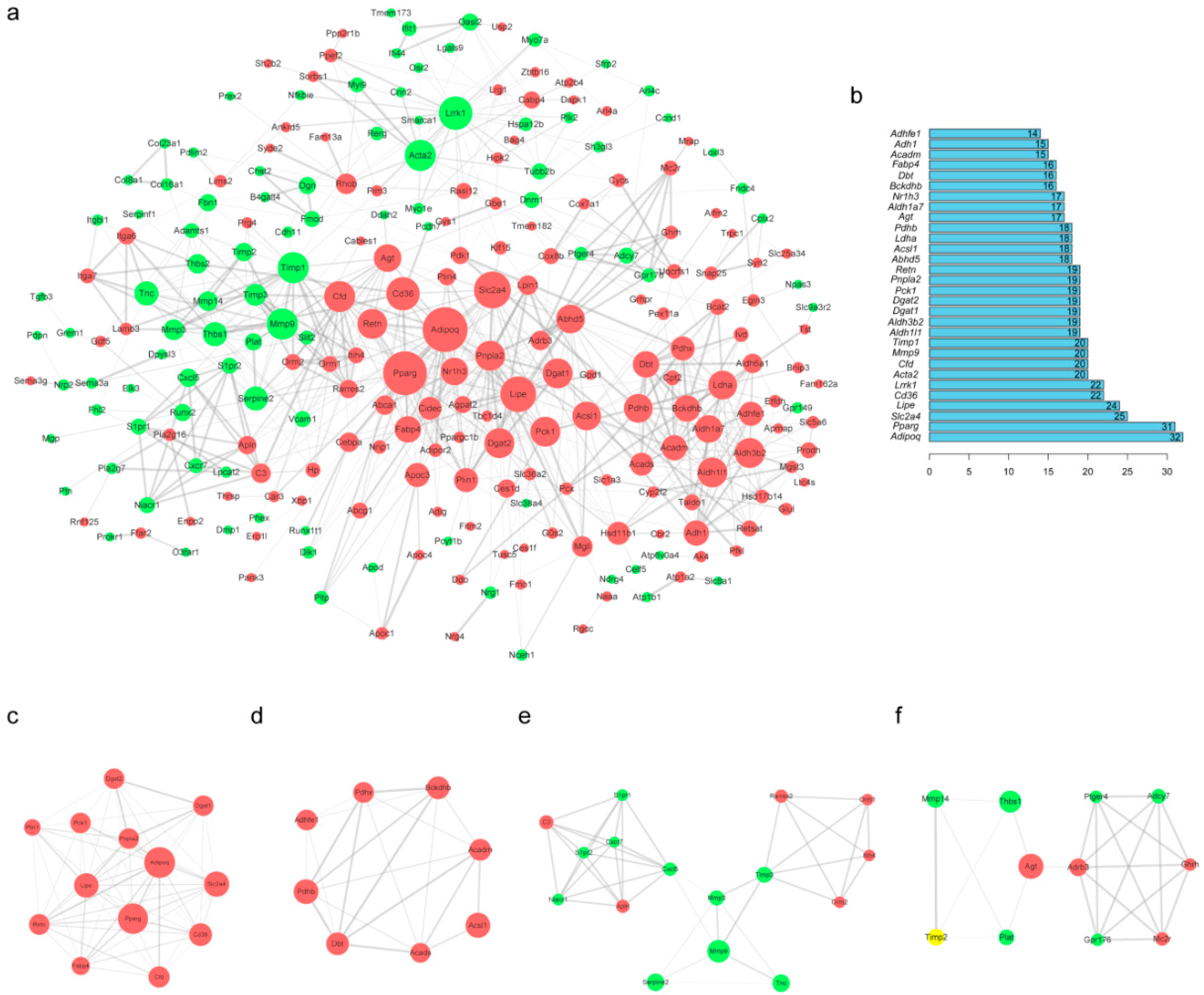
2.5. Protein–Protein Interaction (PPI) Network Construction and Module Analysis of DEGs
2.6. Verification of Changes in DEG Expression During 3T3-L1 Preadipocyte Differentiation
3. Discussions
4. Materials and Methods
4.1. Gene Expression Profile Data
4.2. Identification of DEGs
4.3. Integration of Gene Expression Profile Data
4.4. GO Term and KEGG Pathway Enrichment Analyses
4.5. PPI Network Construction and Module Analysis
4.6. Cell Culture, Differentiation, and Lipid Droplet Staining
4.7. qRT-PCR Validation of Key Genes
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| GEO | Gene expression omnibus |
| DEGs | Differentially expressed genes |
| RRA | Robust rank aggregation |
| GO | Gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
| PPIMCODE | Protein–protein interactionMolecular complex detection |
References
- Green, H.; Meuth, M. An established pre-adipose cell line and its differentiation in culture. Cell 1974, 3, 127–133. [Google Scholar] [CrossRef]
- Green, H.; Kehinde, O. Spontaneous heritable changes leading to increased adipose conversion in 3T3 cells. Cell 1976, 7, 105–113. [Google Scholar] [CrossRef]
- Romero, M.; Sabate-Perez, A.; Francis, V.A.; Castrillon-Rodriguez, I.; Diaz-Ramos, A.; Sanchez-Feutrie, M.; Duran, X.; Palacin, M.; Moreno-Navarrete, J.M.; Gustafson, B.; et al. TP53INP2 regulates adiposity by activating beta-catenin through autophagy-dependent sequestration of GSK3beta. Nat. Cell Biol. 2018, 20, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Siersbaek, R.; Madsen, J.; Javierre, B.M.; Nielsen, R.; Bagge, E.K.; Cairns, J.; Wingett, S.W.; Traynor, S.; Spivakov, M.; Fraser, P.; et al. Dynamic Rewiring of Promoter-Anchored Chromatin Loops during Adipocyte Differentiation. Mol. Cell 2017, 66, 420–435. [Google Scholar] [CrossRef] [PubMed]
- Al, A.H.; Evano, B.; Le Digarcher, A.; Gueydan, C.; Dubois, E.; Parrinello, H.; Dantec, C.; Bouschet, T.; Varrault, A.; Journot, L. A systems-level approach to parental genomic imprinting: The imprinted gene network includes extracellular matrix genes and regulates cell cycle exit and differentiation. Genome Res. 2015, 25, 353–367. [Google Scholar]
- Duteil, D.; Metzger, E.; Willmann, D.; Karagianni, P.; Friedrichs, N.; Greschik, H.; Gunther, T.; Buettner, R.; Talianidis, I.; Metzger, D.; et al. LSD1 promotes oxidative metabolism of white adipose tissue. Nat. Commun. 2014, 5, 4093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikkelsen, T.S.; Xu, Z.; Zhang, X.; Wang, L.; Gimble, J.M.; Lander, E.S.; Rosen, E.D. Comparative epigenomic analysis of murine and human adipogenesis. Cell 2010, 143, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Kolde, R.; Laur, S.; Adler, P.; Vilo, J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics 2012, 28, 573–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Xu, H.; Xiao, T.; Cong, L.; Love, M.I.; Zhang, F.; Irizarry, R.A.; Liu, J.S.; Brown, M.; Liu, X.S. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014, 15, 554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Spalding, K.L.; Arner, E.; Westermark, P.O.; Bernard, S.; Buchholz, B.A.; Bergmann, O.; Blomqvist, L.; Hoffstedt, J.; Naslund, E.; Britton, T.; et al. Dynamics of fat cell turnover in humans. Nature 2008, 453, 783–787. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Cui, Q.; Zhang, X.; Luo, X.; Liu, Y.; Zuo, J.; Peng, Y. Long non-coding RNAs regulation in adipogenesis and lipid metabolism: Emerging insights in obesity. Cell Signal. 2018, 51, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Gregoire, F.M.; Smas, C.M.; Sul, H.S. Understanding adipocyte differentiation. Physiol. Rev. 1998, 78, 783–809. [Google Scholar] [CrossRef] [PubMed]
- Selvarajan, S.; Lund, L.R.; Takeuchi, T.; Craik, C.S.; Werb, Z. A plasma kallikrein-dependent plasminogen cascade required for adipocyte differentiation. Nat. Cell Biol. 2001, 3, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; MacDougald, O.A. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 2006, 7, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Loftus, T.M.; Lane, M.D. Modulating the transcriptional control of adipogenesis. Curr. Opin. Genet. Dev. 1997, 7, 603–608. [Google Scholar] [CrossRef]
- Liu, L.; Zheng, L.D.; Zou, P.; Brooke, J.; Smith, C.; Long, Y.C.; Almeida, F.A.; Liu, D.; Cheng, Z. FoxO1 antagonist suppresses autophagy and lipid droplet growth in adipocytes. Cell Cycle 2016, 15, 2033–2041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, D.E.; Brown, R.; Navarro, J.; Pilch, P.F. Insulin-regulatable tissues express a unique insulin-sensitive glucose transport protein. Nature 1988, 333, 183–185. [Google Scholar] [CrossRef] [PubMed]
- Hertzel, A.V.; Bernlohr, D.A. The mammalian fatty acid-binding protein multigene family: Molecular and genetic insights into function. Trends Endocrinol. Metab. 2000, 11, 175–180. [Google Scholar] [CrossRef]
- Abumrad, N.A.; El-Maghrabi, M.R.; Amri, E.Z.; Lopez, E.; Grimaldi, P.A. Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation. Homology with human CD36. J. Biol. Chem. 1993, 268, 17665–17668. [Google Scholar] [PubMed]
- Cases, S.; Stone, S.J.; Zhou, P.; Yen, E.; Tow, B.; Lardizabal, K.D.; Voelker, T.; Farese, R.J. Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J. Biol. Chem. 2001, 276, 38870–38876. [Google Scholar] [CrossRef] [PubMed]
- Cases, S.; Smith, S.J.; Zheng, Y.W.; Myers, H.M.; Lear, S.R.; Sande, E.; Novak, S.; Collins, C.; Welch, C.B.; Lusis, A.J.; et al. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA 1998, 95, 13018–13023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lardizabal, K.D.; Mai, J.T.; Wagner, N.W.; Wyrick, A.; Voelker, T.; Hawkins, D.J. DGAT2 is a new diacylglycerol acyltransferase gene family: Purification, cloning, and expression in insect cells of two polypeptides from Mortierella ramanniana with diacylglycerol acyltransferase activity. J. Biol. Chem. 2001, 276, 38862–38869. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Kawarabayasi, Y.; Kondo, J.; Abe, T.; Nishikawa, K.; Kimura, S.; Hashimoto, T.; Yamamoto, T. Structure and regulation of rat long-chain acyl-CoA synthetase. J. Biol. Chem. 1990, 265, 8681–8685. [Google Scholar] [PubMed]
- Reilich, P.; Horvath, R.; Krause, S.; Schramm, N.; Turnbull, D.M.; Trenell, M.; Hollingsworth, K.G.; Gorman, G.S.; Hans, V.H.; Reimann, J.; et al. The phenotypic spectrum of neutral lipid storage myopathy due to mutations in the PNPLA2 gene. J. Neurol. 2011, 258, 1987–1997. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Ramakrishnan, G.; Chandramohan, C.; Rajasekharan, R. CGI-58, the causative gene for Chanarin-Dorfman syndrome, mediates acylation of lysophosphatidic acid. J. Biol. Chem. 2008, 283, 24525–24533. [Google Scholar] [CrossRef] [PubMed]
- Song, N.J.; Kim, S.; Jang, B.H.; Chang, S.H.; Yun, U.J.; Park, K.M.; Waki, H.; Li, D.Y.; Tontonoz, P.; Park, K.W. Small Molecule-Induced Complement Factor D (Adipsin) Promotes Lipid Accumulation and Adipocyte Differentiation. PLoS ONE 2016, 11, e162228. [Google Scholar] [CrossRef] [PubMed]
- Meier, U.; Gressner, A.M. Endocrine regulation of energy metabolism: Review of pathobiochemical and clinical chemical aspects of leptin, ghrelin, adiponectin, and resistin. Clin. Chem. 2004, 50, 1511–1525. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Li, Q.; Liu, M.; Ma, W.; Zhou, T.; Xue, C.; Cai, X. Substrate stiffness regulated migration and invasion ability of adenoid cystic carcinoma cells via RhoA/ROCK pathway. Cell. Prolif. 2018, 51, e12442. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, P.E.; Abdelkader, N.F.; El, A.S.; El-Shabrawy, O.A.; Zaki, H.F. Extracellular Matrix Remodeling and Modulation of Inflammation and Oxidative Stress by Sulforaphane in Experimental Diabetic Peripheral Neuropathy. Inflammation 2018, 41, 1460–1476. [Google Scholar] [CrossRef] [PubMed]
- Chavey, C.; Mari, B.; Monthouel, M.N.; Bonnafous, S.; Anglard, P.; Van Obberghen, E.; Tartare-Deckert, S. Matrix metalloproteinases are differentially expressed in adipose tissue during obesity and modulate adipocyte differentiation. J. Biol. Chem. 2003, 278, 11888–11896. [Google Scholar] [CrossRef] [PubMed]
- Bouloumie, A.; Sengenes, C.; Portolan, G.; Galitzky, J.; Lafontan, M. Adipocyte produces matrix metalloproteinases 2 and 9: Involvement in adipose differentiation. Diabetes 2001, 50, 2080–2086. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Baek, S.Y.; Jang, E.J.; Ku, S.K.; Kim, K.M.; Ki, S.H.; Kim, C.E.; Park, K.I.; Kim, S.C.; Kim, Y.W. Oxyresveratrol ameliorates nonalcoholic fatty liver disease by regulating hepatic lipogenesis and fatty acid oxidation through liver kinase B1 and AMP-activated protein kinase. Chem. Biol. Interact. 2018, 289, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.; Tontonoz, P. Liver X receptors in lipid metabolism: Opportunities for drug discovery. Nat. Rev. Drug Discov. 2014, 13, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Beale, E.G.; Hammer, R.E.; Antoine, B.; Forest, C. Disregulated glyceroneogenesis: PCK1 as a candidate diabetes and obesity gene. Trends Endocrinol. Metab. 2004, 15, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.C.; Lane, M.D. The enzymatic carboxylation of phosphoenolpyruvate. II. Purification and properties of liver mitochondrial phosphoenolpyruvate carboxykinase. J. Biol. Chem. 1966, 241, 2413–2420. [Google Scholar] [PubMed]
- Beale, E.G.; Harvey, B.J.; Forest, C. PCK1 and PCK2 as candidate diabetes and obesity genes. Cell Biochem. Biophys. 2007, 48, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, T.; Takagi, S.; Naganuma, T.; Kihara, A. Mouse aldehyde dehydrogenase ALDH3B2 is localized to lipid droplets via two C-terminal tryptophan residues and lipid modification. Biochem. J. 2015, 465, 79–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Mao, X.; Cai, T.; Luo, J.; Wei, L. KOBAS server: A web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006, 34, W720–W724. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Mi, L.; Chen, Y.; Zheng, X.; Li, Y.; Zhang, Q.; Mo, D.; Yang, G. MicroRNA-139-5p Suppresses 3T3-L1 Preadipocyte Differentiation Through Notch and IRS1/PI3K/Akt Insulin Signaling Pathways. J. Cell Biochem. 2015, 116, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
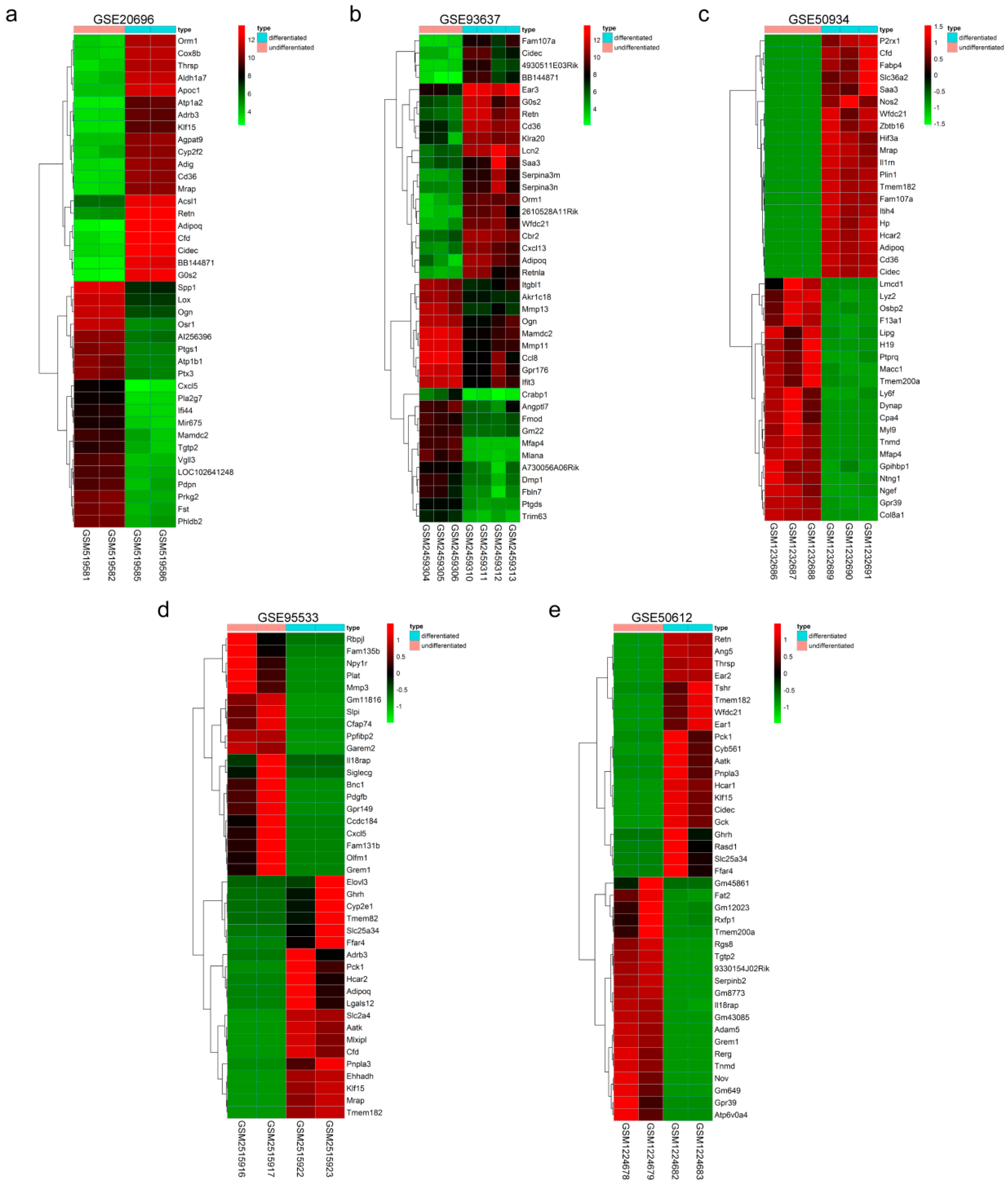

| Reference | GEO | Platform | Study Type | Differentiation Stage of Samples | Number of DEGs (Up/Down) | |
|---|---|---|---|---|---|---|
| Undifferentiated | Differentiated | |||||
| Mikkelsen, T.S. et al. (2010) [7] | GSE20696 | GPL1261 | microarray | GSM519581 GSM519582 | GSM519585 GSM519586 | 2898 (1425/1473) |
| Romero, M. et al. (2018) [3] | GSE93637 | GPL1261 | microarray | GSM2459304 GSM2459305 GSM2459306 | GSM2459310 GSM2459311 GSM2459312 GSM2459313 | 1119 (591/528) |
| Duteil, D. et al. (2014) [6] | GSE50934 | GPL13112 | RNA-seq | GSM1232686 GSM1232687 GSM1232688 | GSM1232689 GSM1232690 GSM1232691 | 2685 (1552/1133) |
| Siersbæk, R. et al. (2017) [4] | GSE95533 | GPL18480 | RNA-seq | GSM2515916 GSM2515917 | GSM2515922 GSM2515923 | 4416 (2197/2219) |
| Al, A.H. et al. (2015) [5] | GSE50612 | GPL13112 | RNA-seq | GSM1224678 GSM1224679 | GSM1224682 GSM1224683 | 3966 (2170/1796) |
| Top 20 Upregulated Genes | Top 20 Downregulated Genes | ||||||
|---|---|---|---|---|---|---|---|
| Symbol | log2FC | p-Value | Corrected p-Value | Symbol | log2FC | p-Value | Corrected p-Value |
| Cidec | 8.815805 | 2.15 × 10−15 | 5.34 × 10−11 | Bnc1 | −4.27787 | 7.38 × 10−12 | 1.83 × 10−7 |
| Retn | 8.50511 | 5.14 × 10−15 | 1.28 × 10−10 | Pcdh18 | −3.56837 | 8.39 × 10−11 | 2.09 × 10−6 |
| G0s2 | 8.359012 | 2.39 × 10−14 | 5.94 × 10−10 | Nrg1 | −3.52592 | 1.30 × 10−10 | 3.24 × 10−6 |
| Adipoq | 9.012189 | 1.81 × 10−13 | 4.51 × 10−9 | Ogn | −3.95531 | 2.77 × 10−10 | 6.89 × 10−6 |
| Cfd | 8.441624 | 1.81 × 10−13 | 4.51 × 10−9 | Mmp9 | −3.64602 | 3.46 × 10−10 | 8.60 × 10−6 |
| Adig | 7.229626 | 1.17 × 10−12 | 2.90 × 10−8 | Tubb2b | −2.96698 | 3.77 × 10−10 | 9.37 × 10−6 |
| Orm1 | 7.280763 | 9.69 × 10−12 | 2.41 × 10−7 | Plat | −3.95792 | 4.36 × 10−10 | 1.09 × 10−5 |
| Dgat2 | 6.066281 | 1.06 × 10−11 | 2.63 × 10−7 | Slit2 | −3.3568 | 7.55 × 10−10 | 1.88 × 10−5 |
| Mc2r | 6.186546 | 1.10 × 10−11 | 2.75 × 10−7 | Rerg | −3.95606 | 1.10 × 10−9 | 2.73 × 10−5 |
| Lgals12 | 6.873706 | 1.67 × 10−11 | 4.15 × 10−7 | Thbs1 | −3.27764 | 1.16 × 10−9 | 2.87 × 10−5 |
| Mrap | 7.563215 | 2.17 × 10−11 | 5.39 × 10−7 | Nfkbie | −2.84309 | 1.82 × 10−9 | 4.53 × 10−5 |
| Acsl1 | 6.016773 | 3.73 × 10−11 | 9.29 × 10−7 | Il17rd | −3.02351 | 2.32 × 10−9 | 5.77 × 10−5 |
| Wfdc21 | 7.632637 | 4.86 × 10−11 | 1.21 × 10−6 | Il1rl1 | −3.70607 | 2.50 × 10−9 | 6.21 × 10−5 |
| Syn2 | 5.030408 | 8.88 × 10−11 | 2.21 × 10−6 | Tnc | −3.78602 | 4.22 × 10−9 | 1.05 × 10−4 |
| Car3 | 5.021359 | 1.14 × 10−10 | 2.83 × 10−6 | Ptger4 | −3.15885 | 4.22 × 10−9 | 1.05 × 10−4 |
| Fabp4 | 6.257684 | 1.34 × 10−10 | 3.32 × 10−6 | Fbn1 | −2.96534 | 4.99 × 10−9 | 1.24 × 10−4 |
| Cd36 | 7.115236 | 1.48 × 10−10 | 3.69 × 10−6 | Atp1b1 | −4.18269 | 5.09 × 10−9 | 1.27 × 10−4 |
| Hp | 6.709448 | 1.60 × 10−10 | 3.98 × 10−6 | Ptprv | −3.41098 | 6.87 × 10−9 | 1.71 × 10−4 |
| Pnpla2 | 5.031089 | 1.77 × 10−10 | 4.40 × 10−6 | Siglecg | −3.88948 | 7.62 × 10−9 | 1.90 × 10−4 |
| Gpd1 | 6.752793 | 2.03 × 10−10 | 5.05 × 10−6 | Pla2g7 | −3.91415 | 7.64 × 10−9 | 1.90 × 10−4 |
| DEGs | Term | Category | Count | FDR |
|---|---|---|---|---|
| up | brown fat cell differentiation | BP | 19 | 1.63 × 10−23 |
| lipid metabolic process | BP | 27 | 3.04 × 10−8 | |
| oxidation-reduction process | BP | 32 | 6.77 × 10−8 | |
| metabolic process | BP | 23 | 3.03 × 10−5 | |
| fat cell differentiation | BP | 10 | 4.76 × 10−4 | |
| lipid storage | BP | 7 | 8.27 × 10−4 | |
| triglyceride catabolic process | BP | 6 | 4.12 × 10−3 | |
| glucose homeostasis | BP | 10 | 3.70 × 10−2 | |
| white fat cell differentiation | BP | 5 | 4.44 × 10−2 | |
| mitochondrion | CC | 52 | 9.65 × 10−8 | |
| lipid particle | CC | 12 | 2.11 × 10−7 | |
| cytosol | CC | 42 | 8.61 × 10−3 | |
| intracellular membrane-bounded organelle | CC | 24 | 1.60 × 10−2 | |
| mitochondrial membrane | CC | 9 | 2.25 × 10−2 | |
| oxidoreductase activity | MF | 27 | 3.98 × 10−6 | |
| down | cell adhesion | BP | 20 | 8.40 × 10−6 |
| positive regulation of cell-substrate adhesion | BP | 7 | 1.19 × 10−3 | |
| positive regulation of neuron projection development | BP | 9 | 1.74 × 10−2 | |
| extracellular region | CC | 51 | 4.64 × 10−14 | |
| proteinaceous extracellular matrix | CC | 24 | 2.78 × 10−13 | |
| extracellular matrix | CC | 21 | 1.28 × 10−10 | |
| extracellular space | CC | 41 | 1.82 × 10−9 | |
| basement membrane | CC | 11 | 4.33 × 10−6 | |
| calcium ion binding | MF | 24 | 4.46 × 10−6 | |
| integrin binding | MF | 9 | 1.34 × 10−3 | |
| heparin binding | MF | 13 | 2.56 × 10−6 |
| DEGs | Term | Count | FDR |
|---|---|---|---|
| up | Metabolic pathways | 42 | 1.34 × 10−16 |
| PPAR signaling pathway | 15 | 3.33 × 10−15 | |
| Regulation of lipolysis in adipocytes | 9 | 1.20 × 10−8 | |
| AMPK signaling pathway | 10 | 5.06 × 10−7 | |
| Valine, leucine and isoleucine degradation | 7 | 3.31 × 10−6 | |
| Non-alcoholic fatty liver disease (NAFLD) | 9 | 2.05 × 10−5 | |
| Propanoate metabolism | 5 | 5.31 × 10−5 | |
| Glycerolipid metabolism | 6 | 5.31 × 10−5 | |
| Metabolism of xenobiotics by cytochrome P450 | 6 | 7.77 × 10−5 | |
| Glycolysis/Gluconeogenesis | 6 | 7.77 × 10−5 | |
| Pyruvate metabolism | 5 | 8.57 × 10−5 | |
| Nitrogen metabolism | 4 | 8.57 × 10−5 | |
| Insulin resistance | 7 | 8.57 × 10−5 | |
| Fat digestion and absorption | 5 | 8.57 × 10−5 | |
| Adipocytokine signaling pathway | 6 | 8.90 × 10−5 | |
| Carbon metabolism | 7 | 1.09 × 10−4 | |
| Proximal tubule bicarbonate reclamation | 4 | 1.63 × 10−4 | |
| Fatty acid degradation | 5 | 1.65 × 10−4 | |
| Glucagon signaling pathway | 6 | 4.20 × 10−4 | |
| Biosynthesis of amino acids | 5 | 1.26 × 10−3 | |
| down | Malaria | 4 | 7.99 × 10−3 |
| Transcriptional misregulation in cancer | 6 | 7.99 × 10−3 | |
| Rheumatoid arthritis | 4 | 2.24 × 10−2 | |
| Bladder cancer | 3 | 2.67 × 10−2 | |
| TNF signaling pathway | 4 | 3.24 × 10−2 | |
| Focal adhesion | 5 | 3.24 × 10−2 | |
| Endocrine and other factor-regulated calcium reabsorption | 3 | 3.24 × 10−2 | |
| Glycosaminoglycan biosynthesis—keratan sulfate | 2 | 3.51 × 10−2 | |
| Synaptic vesicle cycle | 3 | 3.51 × 10−2 | |
| FoxO signaling pathway | 4 | 3.51 × 10−2 | |
| Bile secretion | 3 | 4.35 × 10−2 |
| Method | Term | Count | FDR | Genes |
|---|---|---|---|---|
| GO | metabolic process | 10 | 3.98 × 10−5 | Dbt, Aldh1l1, Acsl1, Acadm, Bckdhb, Aldh3b2, Pnpla2, Aldh1a7, Pdhb, Lipe |
| oxidation-reduction process | 9 | 1.12 × 10−2 | Ldha, Aldh1l1, Acadm, Adhfe1, Adh1, Bckdhb, Aldh3b2, Aldh1a7, Pdhb | |
| lipid storage | 4 | 1.51 × 10−2 | Dgat1, Cd36, Dgat2, Pnpla2 | |
| brown fat cell differentiation | 4 | 3.07 × 10−2 | Slc2a4, Pparg, Fabp4, Adipoq | |
| negative regulation of sequestering of triglyceride | 3 | 3.54 × 10−2 | Pparg, Abhd5, Pnpla2 | |
| KEGG | PPAR signaling pathway | 8 | 2.89 × 10−13 | Acadm, Adipoq, Fabp4, Cd36, Acsl1, Pck1, Pparg, Nr1h3 |
| Metabolic pathways | 13 | 1.61 × 10−10 | Acadm, Adh1, Bckdhb, Dbt, Dgat1, Acsl1, Ldha, Pck1, Aldh1a7, Pnpla2, Dgat2, Pdhb, Aldh3b2 | |
| AMPK signaling pathway | 6 | 1.88 × 10−8 | Adipoq, Cd36, Lipe, Pck1, Pparg, Slc2a4 | |
| Glycolysis/Gluconeogenesis | 5 | 3.90 × 10−8 | Adh1, Ldha, Pck1, Pdhb, Aldh3b2 | |
| Adipocytokine signaling pathway | 5 | 5.04 × 10−8 | Adipoq, Cd36, Acsl1, Pck1, Slc2a4 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Wang, L.; Li, S.; Zhang, W.; Ma, X.; Cheng, G.; Yang, W.; Zan, L. Identification of Potential Key Genes Associated with Adipogenesis through Integrated Analysis of Five Mouse Transcriptome Datasets. Int. J. Mol. Sci. 2018, 19, 3557. https://doi.org/10.3390/ijms19113557
Zhang S, Wang L, Li S, Zhang W, Ma X, Cheng G, Yang W, Zan L. Identification of Potential Key Genes Associated with Adipogenesis through Integrated Analysis of Five Mouse Transcriptome Datasets. International Journal of Molecular Sciences. 2018; 19(11):3557. https://doi.org/10.3390/ijms19113557
Chicago/Turabian StyleZhang, Song, Li Wang, Shijun Li, Wenzhen Zhang, Xueyao Ma, Gong Cheng, Wucai Yang, and Linsen Zan. 2018. "Identification of Potential Key Genes Associated with Adipogenesis through Integrated Analysis of Five Mouse Transcriptome Datasets" International Journal of Molecular Sciences 19, no. 11: 3557. https://doi.org/10.3390/ijms19113557