Thermodynamic Characterization of the Ca2+-Dependent Interaction Between SOUL and ALG-2
Abstract
:1. Introduction
2. Results
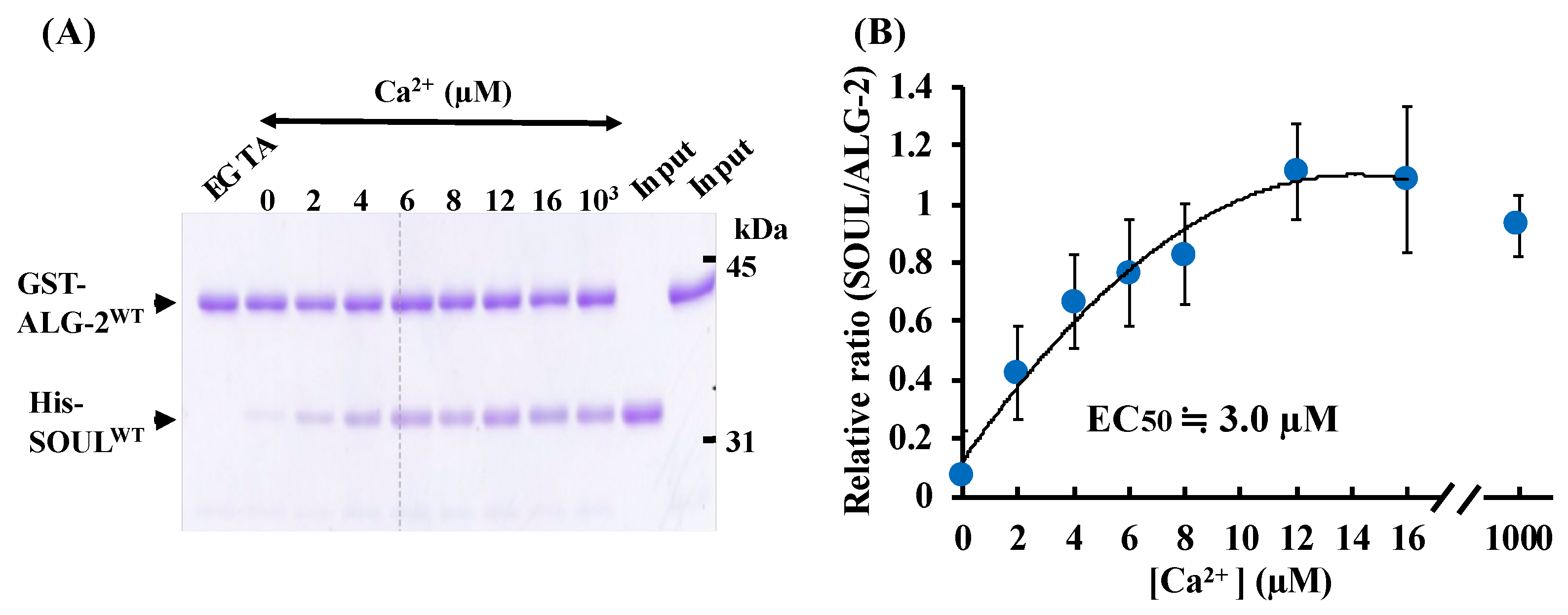
2.1. Ca2+-Dependent Interaction Between SOULWT and ALG-2WT
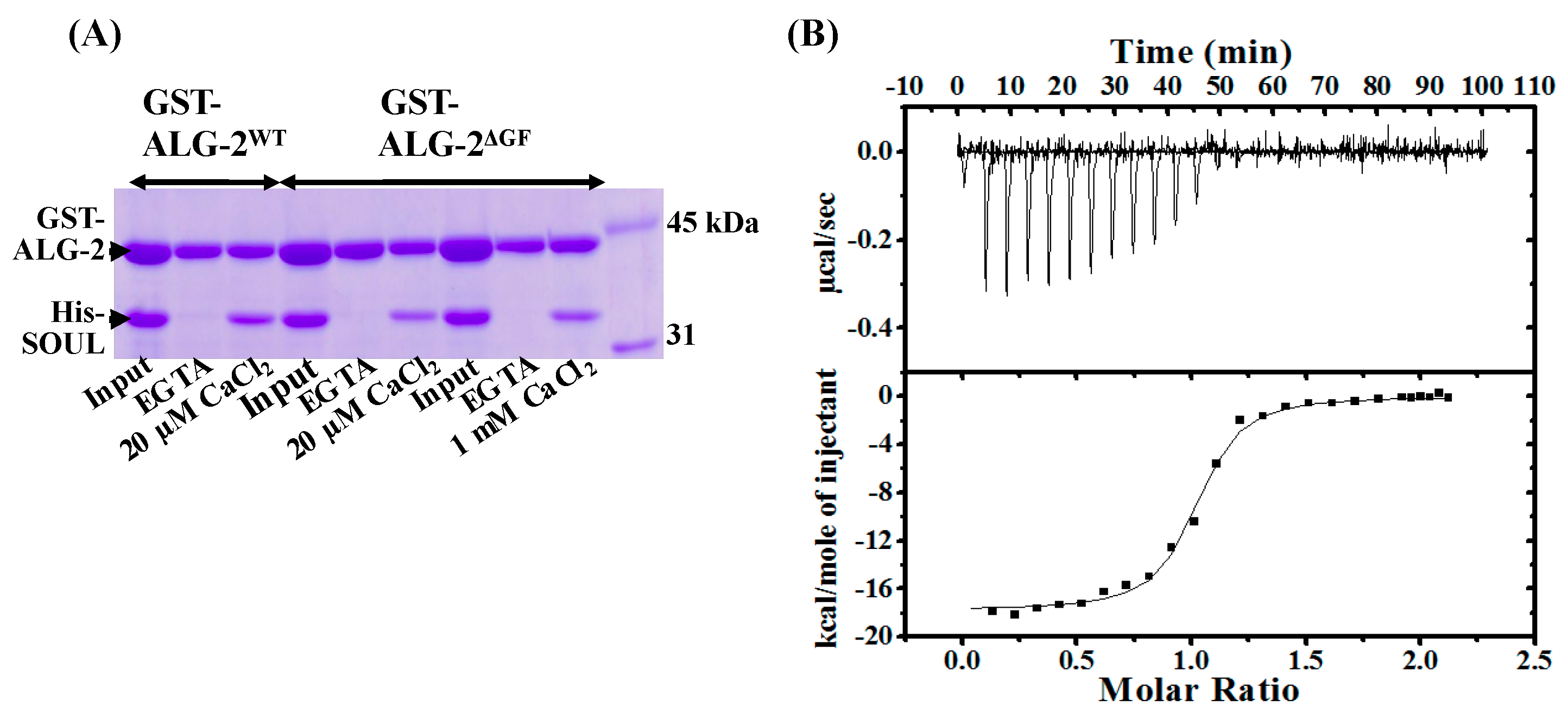
2.2. Interaction of SOULWT with the ALG-2∆GF Variant
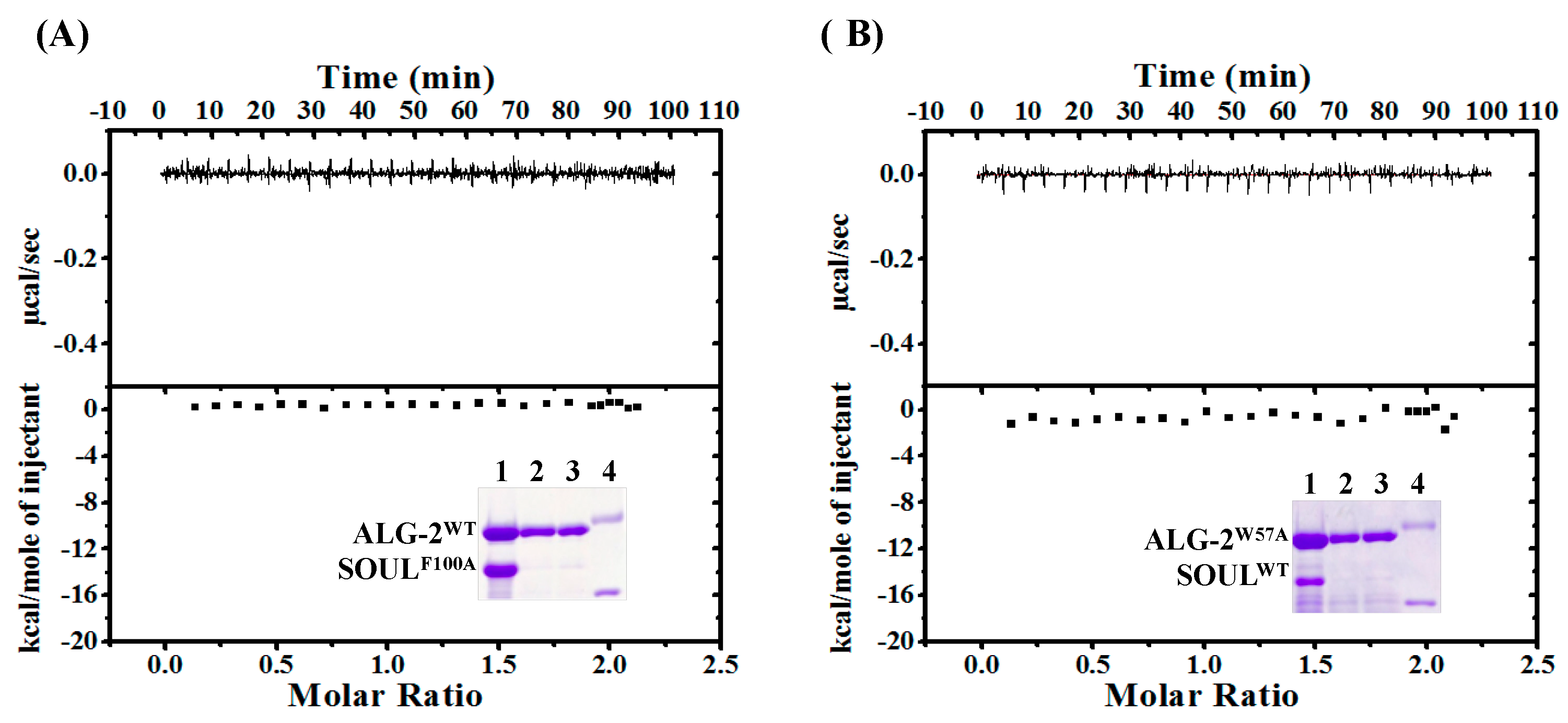
2.3. Effects of the Mutation of SOUL Phe100 and ALG-2 Trp57
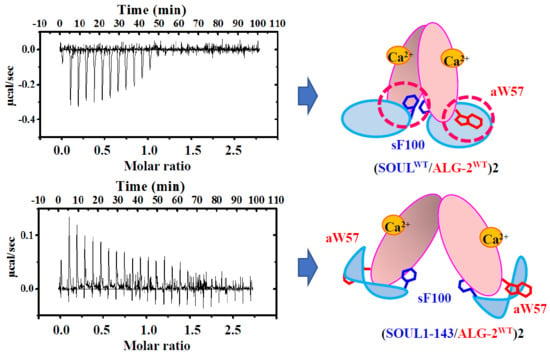
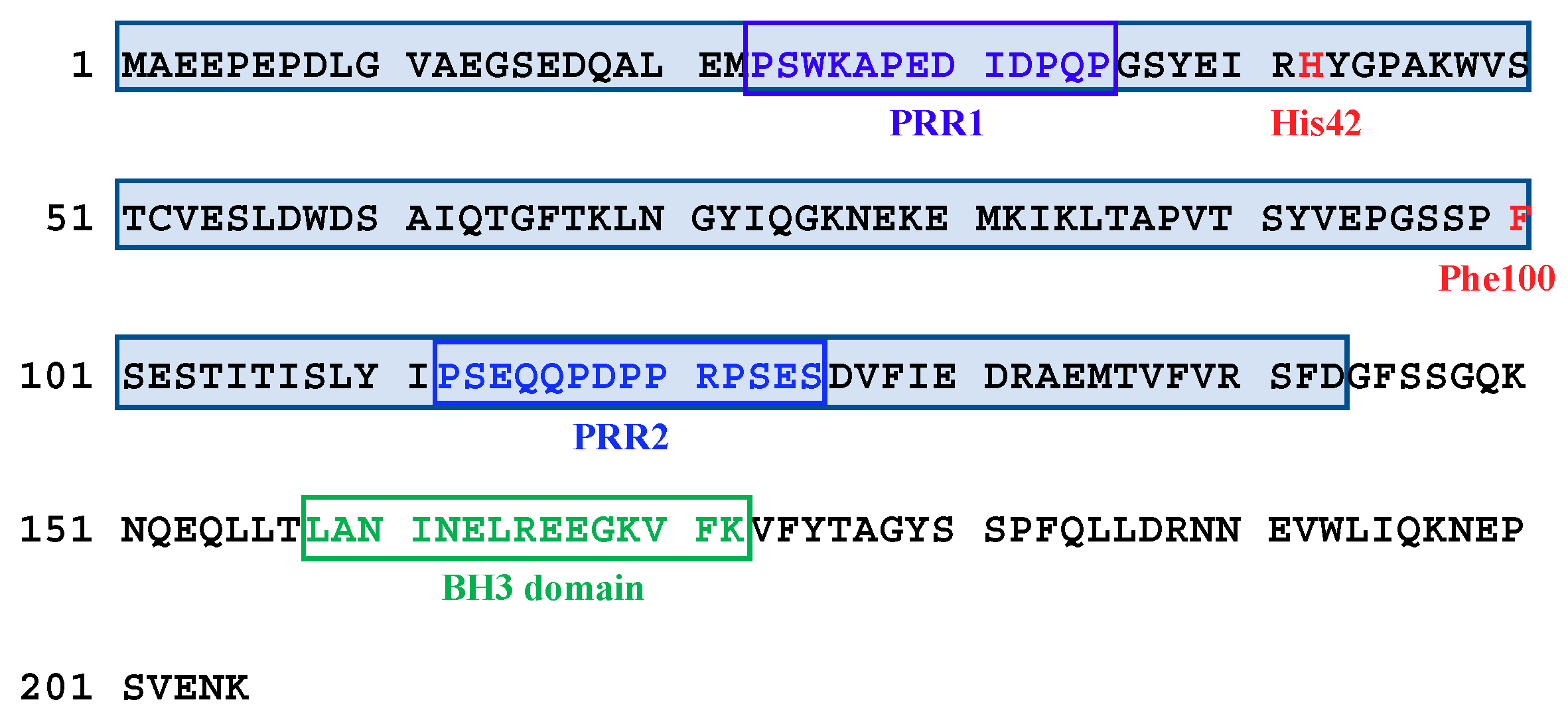
2.4. Interactions of the Truncated SOUL Mutants and ALG-2WT
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction
4.2. Preparation of SOUL and ALG-2 Proteins
4.3. Pulldown Assay
4.4. Gel filtration Analysis
4.5. Isothermal Titration Calorimetry (ITC) Measurements
4.6. Circular Dichroism (CD) Spectra
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABM | ALG-2-binding motif |
| ALIX | ALG-2-interacting protein X |
| ΔCp | Heat capacity change |
| ESCRT | Endosomal sorting complex required for transport |
| HEBP | Heme-binding protein |
| PAGE | Polyacrylamide gel electrophoresis |
| PLSCR3 | Phospholipid scramblase 3 |
| PRR | Proline-rich region |
| TSG101 | Tumor susceptibility gene 101 |
References
- Zylka, M.J.; Reppert, S.M. Discovery of a putative heme-binding protein family (SOUL/HBP) by two-tissue suppression subtractive hybridization and database searches. Brain Res. Mol. 1999, 74, 175–181. [Google Scholar] [CrossRef]
- Fortunato, A.E.; Sordino, P.; Andreakis, N. Evolution of the SOUL heme-binding protein superfamily across Eukarya. J. Mol. Evol. 2016, 82, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Khanna, R.; Shen, Y.; Toledo-Ortiz, G.; Kikis, E.A.; Johannesson, H.; Hwang, Y.S.; Quail, P.H. Functional profiling reveals that only a small number of phytochrome-regulated early-response genes in Arabidopsis are necessary for optimal deetiolation. Plant Cell 2006, 18, 2157–2171. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Mochizuki, N.; Masuda, T.; Buckhout, T.J. Disrupting the bimolecular binding of the haem-binding protein 5 (AtHBP5) to haem oxygenase 1 (HY1) leads to oxidative stress in Arabidopsis. J. Exp. Bot. 2012, 63, 5967–5978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taketani, S.; Adachi, Y.; Kohno, H.; Ikehara, S.; Tokunaga, R.; Ishii, T. Molecular characterization of a newly identified heme-binding protein induced during differentiation of urine erythroleukemia cells. J. Biol. Chem. 1998, 273, 31388–31394. [Google Scholar] [CrossRef] [PubMed]
- Babusiak, M.; Man, P.; Sutak, R.; Petrak, J.; Vyoral, D. Identification of heme binding protein complexes in murine erythroleukemic cells: Study by a novel two-dimensional native separation-liquid chromatography and electrophoresis. Proteomics 2005, 5, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Sato, E.; Sagami, I.; Uchida, T.; Sato, A.; Kitagawa, T.; Igarashi, J.; Shimizu, T. SOUL in mouse eyes is a new hexameric heme-binding protein with characteristic optical absorption, resonance Raman spectral, and heme-binding properties. Biochemistry 2004, 43, 14189–14198. [Google Scholar] [CrossRef] [PubMed]
- Szigeti, A.; Bellyei, S.; Gasz, B.; Boronkai, A.; Hocsak, E.; Minik, O.; Bognar, Z.; Varbiro, G.; Sumegi, B.; Gallyas, F. Induction of necrotic cell death and mitochondrial permeabilization by heme binding protein 2/SOUL. FEBS Lett. 2006, 580, 6447–6454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szigeti, A.; Hocsak, E.; Rapolti, E.; Racz, B.; Boronkai, A.; Pozsgai, E.; Debreceni, B.; Bognar, Z.; Bellyei, S.; Sumegi, B.; et al. Facilitation of mitochondrial outer and inner membrane permeabilization and cell death in oxidative stress by a novel Bcl-2 homology 3 domain protein. J. Biol. Chem. 2010, 285, 2140–2151. [Google Scholar] [CrossRef] [PubMed]
- Ambrosi, E.; Capaldi, S.; Bovi, M.; Saccomani, G.; Perduca, M.; Monaco, H.L. Structural changes in the BH3 domain of SOUL protein upon interaction with the anti-apoptotic protein Bcl-xL. Biochem. J. 2011, 438, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Rual, J.F.; Venkatesan, K.; Hao, T.; Hirozane-Kishikawa, T.; Dricot, A.; Li, N.; Berriz, G.F.; Gibbons, F.D.; Dreze, M.; Ayivi-Guedehoussou, N.; et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature 2005, 437, 1173–1178. [Google Scholar] [CrossRef] [PubMed]
- Rolland, T.; Tasan, M.; Charloteaux, B.; Pevzner, S.J.; Zhong, Q.; Sahni, N.; Yi, S.; Lemmens, I.; Fontanillo, C.; Mosca, R.; et al. A Proteome-Scale Map of the Human Interactome Network. Cell 2014, 159, 1212–1226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Zhang, X.; Feng, Y.; Zhang, H.; Wang, X.; Zheng, Y.; Qiao, W.; Liu, X. Structural and functional study of apoptosis-linked gene-2·heme-binding protein 2 interactions in HIV-1 production. J. Biol. Chem. 2016, 291, 26670–26685. [Google Scholar] [CrossRef] [PubMed]
- Vito, P.; Lacanà, E.; D’Adamio, L. Interfering with apoptosis: Ca2+-binding protein ALG-2 and Alzheimer’s disease gene ALG-3. Science 1996, 271, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Jang, I.K.; Hu, R.; Lacaná, E.; D’Adamio, L.; Gu, H. Apoptosis-linked gene 2-deficient mice exhibit normal T-cell development and function. Mol. Cell. Biol. 2002, 22, 4094–4100. [Google Scholar] [CrossRef] [PubMed]
- Maki, M.; Suzuki, H.; Shibata, H. Structure and function of ALG-2, a penta-EF-hand calcium-dependent adaptor protein. Sci. China Life Sci. 2011, 54, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Maki, M.; Takahara, T.; Shibata, H. Multifaceted roles of ALG-2 in Ca2+-regulated membrane trafficking. Int. J. Mol. Sci. 2016, 17, 1401. [Google Scholar] [CrossRef] [PubMed]
- Okumura, M.; Takahashi, T.; Shibata, H.; Maki, M. Mammalian ESCRT-III-related protein IST1 has a distinctive met-pro repeat sequence that is essential for interaction with ALG-2 in the presence of Ca2+. Biosci. Biotechnol. Biochem. 2013, 77, 1049–1054. [Google Scholar] [CrossRef] [PubMed]
- Vergarajauregui, S.; Martina, J.A.; Puertollano, R. Identification of the penta-EF-hand protein ALG-2 as a Ca2+-dependent interactor of mucolipin-1. J. Biol. Chem. 2009, 284, 36357–36366. [Google Scholar] [CrossRef] [PubMed]
- Tarabykina, S.; Moller, A.L.; Durussel, I.; Cox, J.; Berchtold, M.W. Two forms of the apoptosis-linked protein ALG-2 with different Ca2+ affinities and target recognition. J. Biol. Chem. 2000, 275, 10514–10518. [Google Scholar] [CrossRef] [PubMed]
- Shibata, H.; Suzuki, H.; Kakiuchi, T.; Inuzuka, T.; Yoshida, H.; Mizuno, T.; Maki, M. Identification of Alix-type and non-Alix-type ALG-2-binding sites in human phospholipid scramblase 3: Differential binding to an alternatively spliced isoform and amino acid-substituted mutants. J. Biol. Chem. 2008, 283, 9623–9632. [Google Scholar] [CrossRef] [PubMed]
- Inuzuka, T.; Suzuki, H.; Kawasaki, M.; Shibata, H.; Wakatsuki, S.; Maki, M. Molecular basis for defect in Alix-binding by alternatively spliced isoform of ALG-2 (ALG-2∆GF122) and structural roles of F122 in target recognition. BMC Struct. Biol. 2010, 10. [Google Scholar] [CrossRef]
- Maki, M.; Yamaguchi, K.; Kitaura, Y.; Satoh, H.; Hitomi, K. Calcium-induced exposure of a hydrophobic surface of mouse ALG-2, which is a member of the penta-EF-hand protein family. J. Biochem. 1998, 124, 1170–1177. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, L.; Crabb, J.W.; Cox, J.; Durussel, I.; Walker, T.M.; van Ginkel, P.R.; Bhattacharya, S.; Dellaria, J.M.; Palczewski, K.; Polans, A.S. Ca2+ binding to EF hands 1 and 3 is essential for the interaction of apoptosis-linked gene-2 with Alix/AIP1 in ocular melanoma. Biochemistry 2004, 43, 11175–11186. [Google Scholar] [CrossRef] [PubMed]
- Henzl, M.T.; Frey, B.B.; Wolf, A.J. ALG-2 divalent-ion affinity: Calorimetric analysis of the des23 versions reveals high-affinity site for Mg2+. Biophys. Chem. 2016, 209, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Tarabykina, S.; Hansen, C.; Berchtold, M.; Cygler, M. Structure of apoptosis-linked protein ALG-2: Insights into Ca2+-induced changes in penta-EF-hand proteins. Structure 2001, 9, 267–275. [Google Scholar] [CrossRef]
- Suzuki, H.; Kawasaki, M.; Inuzuka, T.; Okumura, M.; Kakiuchi, T.; Shibata, H.; Wakatsuki, S.; Maki, M. Structural basis for Ca2+-dependent formation of ALG-2/Alix peptide complex: Ca2+/EF3-driven arginine switch mechanism. Structure 2008, 16, 1562–1573. [Google Scholar] [CrossRef] [PubMed]
- Ilari, A.; Fiorillo, A.; Poser, E.; Lalioti, V.S.; Sundell, G.N.; Ivarsson, Y.; Genovese, I.; Colotti, G. Structural basis of Sorcin-mediated calcium-dependent signal transduction. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Satoh, H.; Nakano, Y.; Shibata, H.; Maki, M. The penta-EF-hand domain of ALG-2 interacts with amino-terminal domains of both annexin VII and annexin XI in a Ca2+-dependent manner. Biochim. Biophys. Acta 2002, 1600, 61–67. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Xia, Y.-L.; Ai, S.-M.; Liang, J.; Sang, P.; Ji, X.-L.; Liu, S.-Q. Insights into Protein–Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef] [PubMed]
- Gomez, J.; Hilser, V.J.; Xie, D.; Freire, E. The heat capacity of proteins. Proteins 1995, 22, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Bergqvist, S.; Williams, M.A.; O’Brien, R.; Ladbury, J.E. Heat capacity effects of water molecules and ions at a protein-DNA interface. J. Mol. Biol. 2004, 4, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Oruganti, S.V.; Gopalan, V.; Foster, M.P. Thermodynamics of Coupled Folding in the Interaction of Archaeal RNase P Proteins RPP21 and RPP29. Biochemistry 2012, 51, 926–935. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Kojima, K.; Zhang, W.; Sasaki, K.; Ito, M.; Suzuki, H.; Kawasaki, M.; Wakatsuki, S.; Takahara, T.; Shibata, H.; et al. Structural analysis of the complex between penta-EF-hand ALG-2 protein and Sec31A peptide reveals a novel target recognition mechanism of ALG-2. Int. J. Mol. Sci. 2015, 16, 3677–3699. [Google Scholar] [CrossRef] [PubMed]
- Lo, K.W.; Zhang, Q.; Li, M.; Zhang, M. Apoptosis-linked gene product ALG-2 is a new member of the calpain small subunit subfamily of Ca2+-binding proteins. Biochemistry 1999, 38, 7498–7508. [Google Scholar] [CrossRef] [PubMed]





| ALG-2 Proteins | n | Kd (nM) | ΔH (kcal/mol) | ΔG (kcal/mol) | T∆S (kcal/mol) |
|---|---|---|---|---|---|
| delta3-23ALG-2WT | 1.16 ± 0.02 | 32.4 ± 6.6 | −17.9 ± 0.7 | −10.2 ± 0.2 | −7.6 ± 0.6 |
| delta3-23ALG-2∆GF | 1.00 ± 0.07 | 62.0 ± 13.7 | −19.8 ± 2.0 | −9.8 ± 0.1 | −9.4 ± 1.6 |
| Target Protein (Peptide) | Binding Affinity (Kd) | Method | Reference |
|---|---|---|---|
| SOUL | 32.4 nM | ITC | this work |
| ALIX (799-814: QGPPYPTYPGYPGYSQ) * | ~0.2 µM | SPR | [24] |
| Annexin A7 (4-PGYPPPPGGYP) * | 60 nM, 0.7 µM | SPR | [31] |
| Annexin A11 (4-PGYPPPPGGYP)) * | 40 nM, 0.5 µM | SPR | [31] |
| PLSCR3 ABS-1 peptide (KGYAPSPPPPYPVTPGYPEPA) * ABS-2 peptide (KQVPAPAPGFALFPSPGPVA) * | 40 nM 25 nM | SPR SPR | [21] |
| Sorcin (12-GYYPGG) | 5 µM | SPR | [30] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mikasa, T.; Kugo, M.; Nishimura, S.; Taketani, S.; Ishijima, S.; Sagami, I. Thermodynamic Characterization of the Ca2+-Dependent Interaction Between SOUL and ALG-2. Int. J. Mol. Sci. 2018, 19, 3802. https://doi.org/10.3390/ijms19123802
Mikasa T, Kugo M, Nishimura S, Taketani S, Ishijima S, Sagami I. Thermodynamic Characterization of the Ca2+-Dependent Interaction Between SOUL and ALG-2. International Journal of Molecular Sciences. 2018; 19(12):3802. https://doi.org/10.3390/ijms19123802
Chicago/Turabian StyleMikasa, Taisuke, Masami Kugo, Seigo Nishimura, Sigeru Taketani, Sumio Ishijima, and Ikuko Sagami. 2018. "Thermodynamic Characterization of the Ca2+-Dependent Interaction Between SOUL and ALG-2" International Journal of Molecular Sciences 19, no. 12: 3802. https://doi.org/10.3390/ijms19123802