Local and Systemic Cytokine Profiling for Pancreatic Ductal Adenocarcinoma to Study Cancer Cachexia in an Era of Precision Medicine
Abstract
:1. Introduction
2. Results
2.1. Xenograft Selection
2.2. PDX Model Preserves Cancer-Associated Cachexia
2.3. Atrophy Related Genes Are Expressed in PDX Models
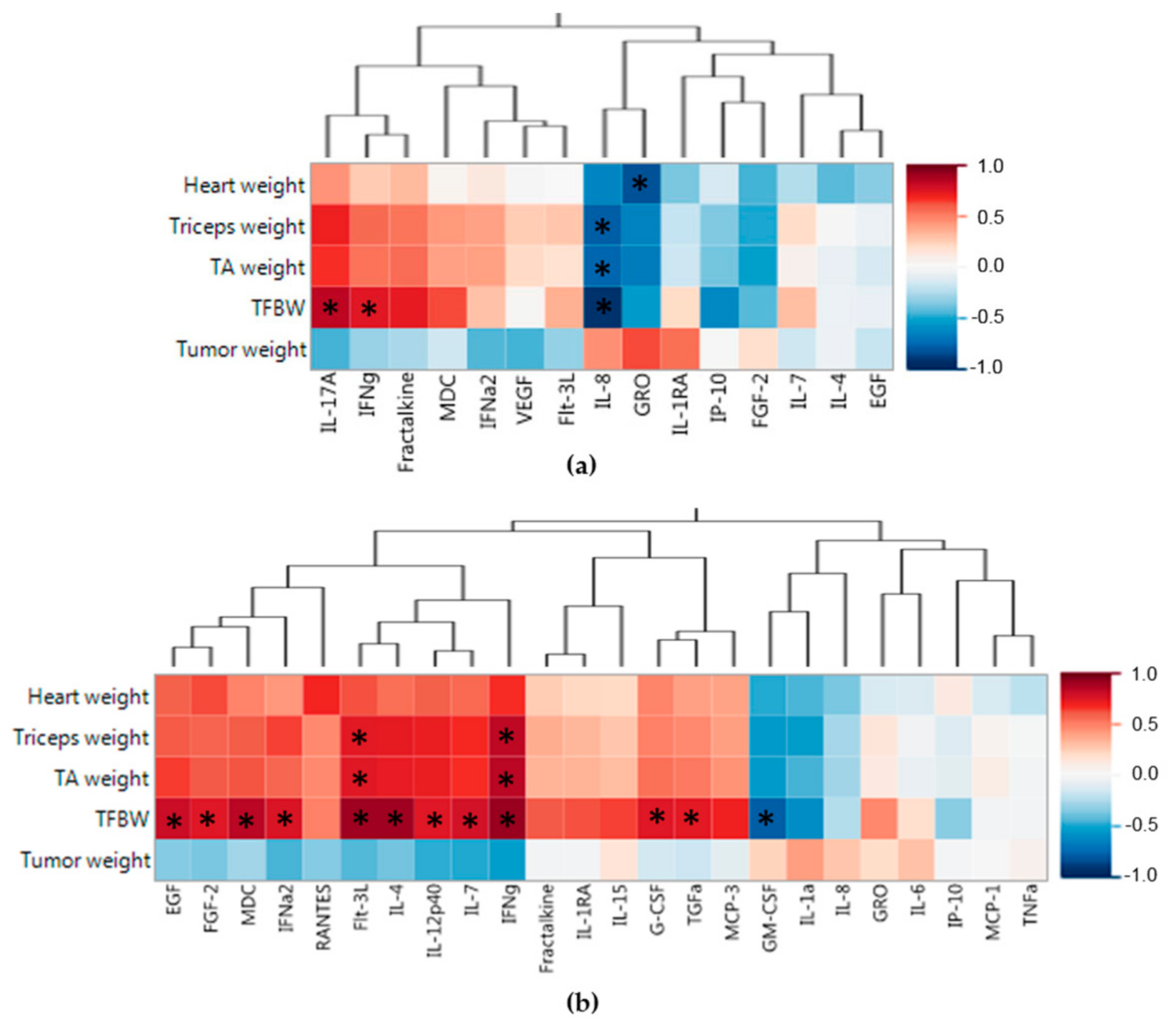
2.4. PDX Model Confers Unique Splenic Soluble Protein Profiles
2.5. PDX Model Confers Unique Tumor Microenvironment Soluble Protein Profiles
2.6. Splenic and Tumor Soluble Protein Associations
3. Discussion
4. Materials and Methods
4.1. Patient Derived Xenograft Model
4.2. Mouse Tissue Harvest and Processing
4.3. Genetic Expression Profiling
4.4. Soluble Protein Analysis
4.5. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PDX | Patient Derived Xenograft |
| PDAC | Pancreatic Ductal Adenocarcinoma |
| NSG | NOD.Cg-Prkdcscid IL2rgtm1Wjl/SzJ |
| IACUC | Institutional Animal Care and Use Committee |
| TA | Tibialis Anterior |
References
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Laviano, A.; Meguid, M.M.; Inui, A.; Muscaritoli, M.; Rossi-Fanelli, F. Therapy insight: Cancer anorexia-cachexia syndrome—When all you can eat is yourself. Nat. Clin. Pract. Oncol. 2005, 2, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Wigmore, S.J.; Plester, C.E.; Richardson, R.A.; Fearon, K.C. Changes in nutritional status associated with unresectable pancreatic cancer. Br. J. Cancer 1997, 75, 106–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baracos, V.E.; Martin, L.; Korc, M.; Guttridge, D.C.; Fearon, K.C.H. Cancer-associated cachexia. Nat. Rev. Dis. Primers 2018, 4, 17105. [Google Scholar] [CrossRef] [PubMed]
- Tisdale, M.J. Mechanisms of cancer cachexia. Physiol. Rev. 2009, 89, 381–410. [Google Scholar] [CrossRef] [PubMed]
- Tisdale, M.J. Cancer cachexia. Curr. Opin. Gastroenterol. 2010, 26, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Couch, M.; Lai, V.; Cannon, T.; Guttridge, D.; Zanation, A.; George, J.; Hayes, D.N.; Zeisel, S.; Shores, C. Cancer cachexia syndrome in head and neck cancer patients: Part I. Diagnosis, impact on quality of life and survival, and treatment. Head Neck 2007, 29, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Delitto, D.; Judge, S.M.; George, T.J., Jr.; Sarosi, G.A.; Thomas, R.M.; Behrns, K.E.; Hughes, S.J.; Judge, A.R.; Trevino, J.G. A clinically applicable muscular index predicts long-term survival in resectable pancreatic cancer. Surgery 2017, 161, 930–938. [Google Scholar] [CrossRef] [PubMed]
- De Lerma Barbaro, A. The complex liaison between cachexia and tumor burden (Review). Oncol. Rep. 2015, 34, 1635–1649. [Google Scholar] [CrossRef] [PubMed]
- Fearon, K.C. The Sir David Cuthbertson Medal Lecture 1991. The mechanisms and treatment of weight loss in cancer. Proc. Nutr. Soc. 1992, 51, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Nixon, D.W.; Lawson, D.H.; Kutner, M.; Ansley, J.; Schwarz, M.; Heymsfield, S.; Chawla, R.; Cartwright, T.H.; Rudman, D. Hyperalimentation of the cancer patient with protein-calorie undernutrition. Cancer Res. 1981, 41, 2038–2045. [Google Scholar] [PubMed]
- Fearon, K.C. Cancer cachexia: Developing multimodal therapy for a multidimensional problem. Eur. J. Cancer 2008, 44, 1124–1132. [Google Scholar] [CrossRef] [PubMed]
- Streat, S.J.; Beddoe, A.H.; Hill, G.L. Aggressive nutritional support does not prevent protein loss despite fat gain in septic intensive care patients. J. Trauma 1987, 27, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Delitto, D.; Judge, S.M.; Delitto, A.E.; Nosacka, R.L.; Rocha, F.G.; DiVita, B.B.; Gerber, M.H.; George, T.J., Jr.; Behrns, K.E.; Hughes, S.J.; et al. Human pancreatic cancer xenografts recapitulate key aspects of cancer cachexia. Oncotarget 2017, 8, 1177–1189. [Google Scholar] [CrossRef] [PubMed]
- Greco, S.H.; Tomkotter, L.; Vahle, A.K.; Rokosh, R.; Avanzi, A.; Mahmood, S.K.; Deutsch, M.; Alothman, S.; Alqunaibit, D.; Ochi, A.; et al. TGF-β Blockade Reduces Mortality and Metabolic Changes in a Validated Murine Model of Pancreatic Cancer Cachexia. PLoS ONE 2015, 10, e0132786. [Google Scholar] [CrossRef] [PubMed]
- Haugen, F.; Labori, K.J.; Noreng, H.J.; Buanes, T.; Iversen, P.O.; Drevon, C.A. Altered expression of genes in adipose tissues associated with reduced fat mass in patients with pancreatic cancer. Arch. Physiol. Biochem. 2011, 117, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Holmer, R.; Goumas, F.A.; Waetzig, G.H.; Rose-John, S.; Kalthoff, H. Interleukin-6: A villain in the drama of pancreatic cancer development and progression. Hepatobiliary Pancreat. Dis. Int. 2014, 13, 371–380. [Google Scholar] [CrossRef]
- Luster, A.D.; Unkeless, J.C.; Ravetch, J.V. Gamma-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature 1985, 315, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Wigmore, S.J.; Fearon, K.C.; Maingay, J.P.; Lai, P.B.; Ross, J.A. Interleukin-8 can mediate acute-phase protein production by isolated human hepatocytes. Am. J. Physiol. 1997, 273, E720–E726. [Google Scholar] [CrossRef] [PubMed]
- Watchorn, T.M.; Dowidar, N.; Dejong, C.H.; Waddell, I.D.; Garden, O.J.; Ross, J.A. The cachectic mediator proteolysis inducing factor activates NF-κB and STAT3 in human Kupffer cells and monocytes. Int. J. Oncol. 2005, 27, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Krzystek-Korpacka, M.; Matusiewicz, M.; Diakowska, D.; Grabowski, K.; Blachut, K.; Kustrzeba-Wojcicka, I.; Banas, T. Impact of weight loss on circulating IL-1, IL-6, IL-8, TNF-α, VEGF-A, VEGF-C and midkine in gastroesophageal cancer patients. Clin. Biochem. 2007, 40, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, K.; Gorgens, S.W.; Raschke, S.; Eckel, J. Myokines in insulin resistance and type 2 diabetes. Diabetologia 2014, 57, 1087–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoyama, T.; Inokuchi, S.; Brenner, D.A.; Seki, E. CX3CL1-CX3CR1 interaction prevents carbon tetrachloride-induced liver inflammation and fibrosis in mice. Hepatology 2010, 52, 1390–1400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prokopchuk, O.; Steinacker, J.M.; Nitsche, U.; Otto, S.; Bachmann, J.; Schubert, E.C.; Friess, H.; Martignoni, M.E. IL-4 mRNA Is Downregulated in the Liver of Pancreatic Cancer Patients Suffering from Cachexia. Nutr. Cancer 2017, 69, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, L.; Olsen, C.H.; Pedersen, B.K.; Hojman, P. Muscle-derived expression of the chemokine CXCL1 attenuates diet-induced obesity and improves fatty acid oxidation in the muscle. Am. J. Physiol. Endocrinol. Metab. 2012, 302, E831–E840. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, L.; Pilegaard, H.; Hansen, J.; Brandt, C.; Adser, H.; Hidalgo, J.; Olesen, J.; Pedersen, B.K.; Hojman, P. Exercise-induced liver chemokine CXCL-1 expression is linked to muscle-derived interleukin-6 expression. J. Physiol. 2011, 589, 1409–1420. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, D.; Cassol, E. Role of cellular metabolism in regulating type I interferon responses: Implications for tumour immunology and treatment. Cancer Lett. 2017, 409, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Heidari, Z.; Bickerdike, R.; Tinsley, J.; Zou, J.; Wang, T.Y.; Chen, T.Y.; Martin, S.A. Regulatory factors controlling muscle mass: Competition between innate immune function and anabolic signals in regulation of Atrogin-1 in Atlantic salmon. Mol. Immunol. 2015, 67, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Bassukas, I.D.; Hofmockel, G.; Maurer-Schultze, B. Treatment with tumor necrosis factor α and interferon α of a human kidney cancer xenograft in nude mice: Evidence for an anticachectic effect of interferon α. Anticancer Res. 1994, 14, 237–245. [Google Scholar] [PubMed]
- Wang, Y.; Zhou, Y.; Graves, D.T. FOXO transcription factors: Their clinical significance and regulation. Biomed. Res. Int. 2014, 2014, 925350. [Google Scholar] [CrossRef] [PubMed]
- Gumucio, J.P.; Mendias, C.L. Atrogin-1, MuRF-1, and sarcopenia. Endocrine 2013, 43, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Bonetto, A.; Aydogdu, T.; Kunzevitzky, N.; Guttridge, D.C.; Khuri, S.; Koniaris, L.G.; Zimmers, T.A. STAT3 activation in skeletal muscle links muscle wasting and the acute phase response in cancer cachexia. PLoS ONE 2011, 6, e22538. [Google Scholar] [CrossRef] [PubMed]
- Rios, R.; Carneiro, I.; Arce, V.M.; Devesa, J. Myostatin is an inhibitor of myogenic differentiation. Am. J. Physiol.-Cell Physiol. 2002, 282, C993–C999. [Google Scholar] [CrossRef] [PubMed]
- Tisdale, M.J. Molecular pathways leading to cancer cachexia. Physiology 2005, 20, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Pfitzenmaier, J.; Vessella, R.; Higano, C.S.; Noteboom, J.L.; Wallace, D., Jr.; Corey, E. Elevation of cytokine levels in cachectic patients with prostate carcinoma. Cancer 2003, 97, 1211–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, B.; Zhang, D.; Wang, S.; Zheng, H.; Wang, X. Association of interleukin-8 with cachexia from patients with low-third gastric cancer. Comp. Funct. Genomics 2009, 212345. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, B.; Tucker, S.L.; Li, D.; Abbruzzese, J.L.; Kurzrock, R. Cytokines in pancreatic carcinoma: Correlation with phenotypic characteristics and prognosis. Cancer 2004, 101, 2727–2736. [Google Scholar] [CrossRef] [PubMed]




| Patient Demographics | G59 | G68 |
|---|---|---|
| Age | 73 | 64 |
| Charlson comorbidity index | 5 | 4 |
| Race | White | White |
| Sex | Female | Female |
| BMI | 27.3 | 29.2 |
| Weight at time of surgery (pounds) | 149 | 170 |
| Weight loss prior to surgery (pounds) | 10 (6.3%) | 25 (12.8%) |
| Psoas Index at time of surgery | 0.639 | 0.526 |
| Neoadjuvant therapy | No | No |
| Operative & Oncologic Factors | ||
| Pathologic diagnosis | Adenocarcinoma | Adenocarcinoma |
| EBL | 600 | 200 |
| Transfusion, intra-op | No | No |
| Transfusion, post-op | No | Yes |
| Portal vein resection | No | No |
| Total lymph nodes | 15 | 18 |
| Positive lymph nodes | 4 | 2 |
| Tumor size (cm) | Indeterminate | 3.8 |
| Margins | Uninvolved | Uninvolved |
| Differentiation | Moderate | Moderate |
| Stage | T3N1 | T3N1 |
| Postoperative Complications | ||
| Length of stay (days) | 7 | 74 |
| Pancreatic leak | No | Yes |
| Post pancreatectomy hemorrhage | No | Yes |
| Major morbidity (clavien III/IV) | No | Yes |
| Death (Postoperative day) | 711 | 169 |
| Splenic Lysate Proteins | Control (n = 9) | G59 (n = 5) | G68 (n = 5) | Control vs. G59 | Control vs. G68 | G59 vs. G68 |
|---|---|---|---|---|---|---|
| Mean (SD) | Mean (SD) | Mean (SD) | P-Value | P-Value | P-Value | |
| FGF-2 | 2587.5 (557.5) | 1491.0 (398.6) | 1690.2 (552.9) | 0.002 | 0.013 | 0.532 |
| VEGF | 75.1 (12.0) | 9.7 (7.7) | 8.9 (3.8) | <0.001 | <0.001 | 0.846 |
| Fractalkine | 56.6 (28.8) | 67.8 (24.3) | 19.8 (13.7) | 0.478 | 0.021 | 0.005 |
| IL-8 | 1.5 (1.1) | 17.4 (15.2) | 155.5 (113.9) | 0.007 | 0.001 | 0.028 |
| GRO | 1.4 (1.2) | 108.2 (51.0) | 158.6 (66.1) | <0.001 | <0.001 | 0.214 |
| MDC | 21.7 (14.9) | 25.7 (7.9) | 3.0 (1.4) | 0.590 | 0.017 | <0.001 |
| IFNα2 | 15.3 (3.7) | 11.8 (2.5) | 10.7 (2.6) | 0.080 | 0.028 | 0.511 |
| IL-7 | 9.8 (1.9) | 8.4 (1.8) | 5.7 (1.6) | 0.229 | 0.002 | 0.031 |
| Flt-3L | 6.4 (3.5) | 3.5 (1.1) | 2.0 (0.7) | 0.107 | 0.019 | 0.034 |
| EGF | 4.6 (1.5) | 2.8 (1.3) | 2.5 (0.9) | 0.040 | 0.013 | 0.667 |
| IFNγ | 4.3 (2.2) | 4.8 (1.6) | 0.8 (0.3) | 0.663 | 0.004 | 0.001 |
| IP-10 | 2.7 (1.7) | 1.2 (0.9) | 4.4 (3.0) | 0.090 | 0.183 | 0.049 |
| IL-17A | 2.4 (0.8) | 2.0 (0.6) | 0.2 (0.3) | 0.308 | <0.001 | <0.001 |
| IL-4 | 1.1 (1.1) | 2.7 (1.1) | 2.3 (0.7) | 0.024 | 0.053 | 0.498 |
| IL-1RA | 0.1 (0.3) | 20.4 (26.3) | 1.4 (0.6) | 0.033 | <0.001 | 0.144 |
| G59 (n = 5) | G68 (n = 5) | ||
|---|---|---|---|
| Mean (SD) | Mean (SD) | P-value | |
| IL-1RA | 1056.9 (198.8) | 248.9 (126.9) | <0.001 |
| GRO | 689.7 (215.8) | 293.5 (185.9) | 0.014 |
| FGF-2 | 635.8 (359.7) | 236.2 (135.2) | 0.049 |
| IL-8 | 194.3 (122.0) | 267.8 (153.8) | 0.426 |
| Fractalkine | 121.8 (60.7) | 62.9 (12.7) | 0.066 |
| RANTES | 88.6 (46.4) | 82.7 (75.6) | 0.885 |
| IP-10 | 13.1 (6.1) | 65.0 (53.0) | 0.061 |
| GM-CSF | 3.6 (1.5) | 16.3 (7.5) | 0.006 |
| TGFα | 15.2 (5.2) | 3.6 (2.1) | 0.002 |
| IL-1α | 4.1 (2.9) | 11.6 (8.1) | 0.085 |
| MDC | 7.0 (2.8) | 2.4 (1.0) | 0.009 |
| TNFα | 2.9 (1.3) | 6.0 (7.2) | 0.364 |
| IFNα2 | 6.6 (1.3) | 2.2 (1.4) | 0.001 |
| IL-15 | 4.9 (0.6) | 3.8 (0.5) | 0.008 |
| MCP-1 | 3.4 (3.9) | 4.8 (6.5) | 0.679 |
| IL-7 | 3.5 (1.5) | 1.5 (0.6) | 0.026 |
| IL-6 | 2.9 (2.2) | 1.3 (1.0) | 0.167 |
| G-CSF | 2.8 (0.6) | 0.8 (0.4) | <0.001 |
| Flt-3L | 2.4 (0.5) | 0.9 (0.3) | <0.001 |
| IFNγ | 2.0 (1.0) | 0.7 (0.4) | 0.022 |
| IL-4 | 1.9 (1.1) | 0.5 (0.1) | 0.020 |
| EGF | 1.6 (1.0) | 0.5 (0.2) | 0.033 |
| MCP-3 | 1.3 (0.9) | 0.5 (0.2) | 0.083 |
| IL-12p40 | 1.2 (0.4) | 0.6 (0.4) | 0.049 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gerber, M.H.; Underwood, P.W.; Judge, S.M.; Delitto, D.; Delitto, A.E.; Nosacka, R.L.; DiVita, B.B.; Thomas, R.M.; Permuth, J.B.; Hughes, S.J.; et al. Local and Systemic Cytokine Profiling for Pancreatic Ductal Adenocarcinoma to Study Cancer Cachexia in an Era of Precision Medicine. Int. J. Mol. Sci. 2018, 19, 3836. https://doi.org/10.3390/ijms19123836
Gerber MH, Underwood PW, Judge SM, Delitto D, Delitto AE, Nosacka RL, DiVita BB, Thomas RM, Permuth JB, Hughes SJ, et al. Local and Systemic Cytokine Profiling for Pancreatic Ductal Adenocarcinoma to Study Cancer Cachexia in an Era of Precision Medicine. International Journal of Molecular Sciences. 2018; 19(12):3836. https://doi.org/10.3390/ijms19123836
Chicago/Turabian StyleGerber, Michael H., Patrick W. Underwood, Sarah M. Judge, Daniel Delitto, Andrea E. Delitto, Rachel L. Nosacka, Bayli B. DiVita, Ryan M. Thomas, Jennifer B. Permuth, Steven J. Hughes, and et al. 2018. "Local and Systemic Cytokine Profiling for Pancreatic Ductal Adenocarcinoma to Study Cancer Cachexia in an Era of Precision Medicine" International Journal of Molecular Sciences 19, no. 12: 3836. https://doi.org/10.3390/ijms19123836