The miRNA Expression Profile of Experimental Autoimmune Encephalomyelitis Reveals Novel Potential Disease Biomarkers
Abstract
:1. Introduction
2. Results
2.1. The MOG35–55-Induced miRNA Expression Profile of Mice with EAE
2.2. Selection of miRNAs and Their Validation by qRT-PCR and Serum miRNA Testing
2.3. Circulating miRNAs in Sera of EAE Mice
2.4. The mRNA Targets of Select miRNAs Identified in EAE
2.5. The mRNA Targets of Select 3 Top miRNAs
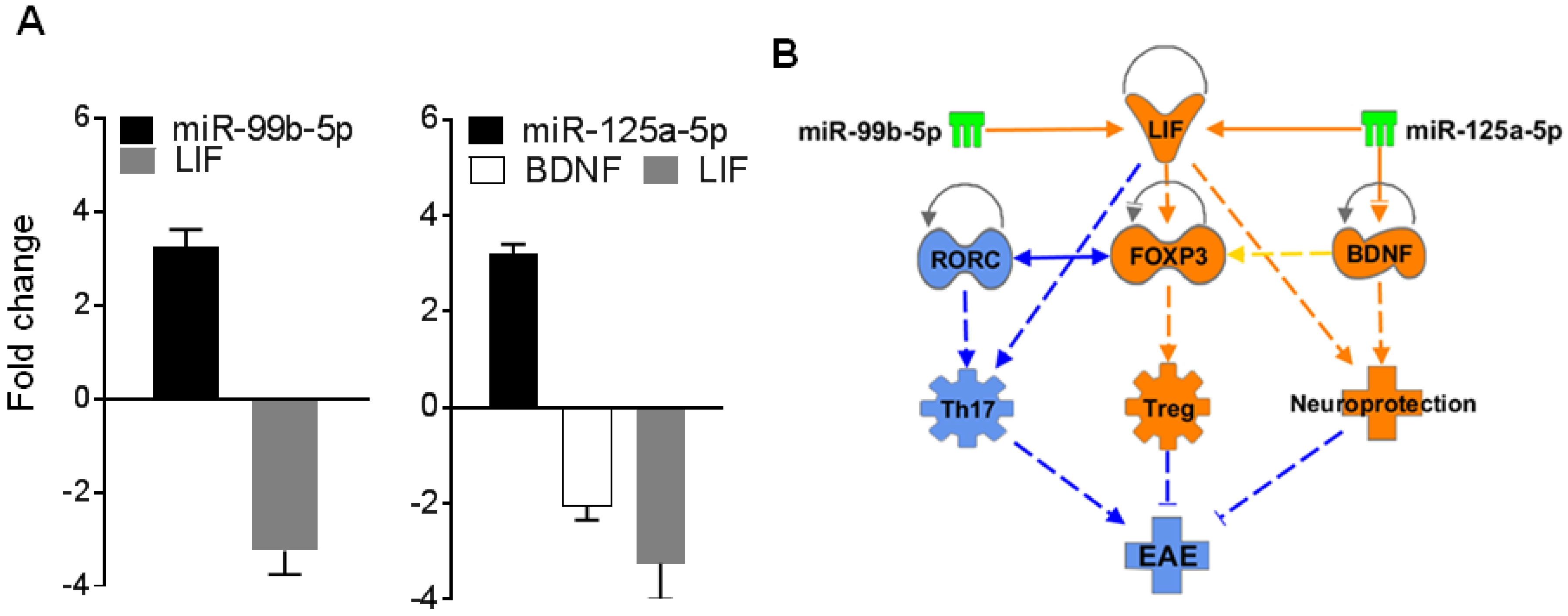
2.6. BDNF and/or LIF Represent Key Targets of miR-125a and 99b
3. Discussion
4. Materials and Methods
4.1. Induction of Experimental Autoimmune Encephalomyelitis (EAE) in Mice
4.2. The Testing of Splenocytes (SPCs) of EAE and Control Mice
4.3. Preparation of Total RNA and Microarray Hybridization for miRNA Expression Analysis
4.4. Micro-RNA Expression Analysis
4.5. Validation of Mature miRNA by Quantitative Real-Time PCR (qRT-PCR)
4.6. Testing miRNA Levels in Sera of Mice
4.7. Ingenuity Pathway Analysis (IPA) for Gene (mRNA) Target Prediction and Network Analysis
4.8. Measurement of mRNA Expression Using qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yadav, S.K.; Mindur, J.E.; Ito, K.; Dhib-Jalbut, S. Advances in the immunopathogenesis of multiple sclerosis. Curr. Opin. Neurol. 2015, 28, 206–219. [Google Scholar] [CrossRef] [PubMed]
- Field, J.; Browning, S.R.; Johnson, L.J.; Danoy, P.; Varney, M.D.; Tait, B.D.; Gandhi, K.S.; Charlesworth, J.C.; Heard, R.N.; The Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene); et al. A polymorphism in the HLA-DPB1 gene is associated with susceptibility to multiple sclerosis. PLoS ONE 2010, 5, e13454. [Google Scholar] [CrossRef] [PubMed]
- Goris, A.; Pauwels, I.; Dubois, B. Progress in multiple sclerosis genetics. Curr. Genom. 2012, 13, 646–663. [Google Scholar] [CrossRef] [PubMed]
- Handel, A.E.; Williamson, A.J.; Disanto, G.; Dobson, R.; Giovannoni, G.; Ramagopalan, S.V. Smoking and multiple sclerosis: An updated meta-analysis. PLoS ONE 2011, 6, e16149. [Google Scholar] [CrossRef] [PubMed]
- Handel, A.E.; Williamson, A.J.; Disanto, G.; Handunnetthi, L.; Giovannoni, G.; Ramagopalan, S.V. An updated meta-analysis of risk of multiple sclerosis following infectious mononucleosis. PLoS ONE 2010, 5, e12496. [Google Scholar] [CrossRef] [PubMed]
- Browne, P.; Chandraratna, D.; Angood, C.; Tremlett, H.; Baker, C.; Taylor, B.V.; Thompson, A.J. Atlas of Multiple Sclerosis 2013: A growing global problem with widespread inequity. Neurology 2014, 83, 1022–1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, C.; Beland, S.G.; Kulaga, S.; Wolfson, C.; Kingwell, E.; Marriott, J.; Koch, M.; Makhani, N.; Morrow, S.; Fisk, J.; et al. Incidence and prevalence of multiple sclerosis in the Americas: A systematic review. Neuroepidemiology 2013, 40, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Kingwell, E.; Marriott, J.J.; Jette, N.; Pringsheim, T.; Makhani, N.; Morrow, S.A.; Fisk, J.D.; Evans, C.; Beland, S.G.; Kulaga, S.; et al. Incidence and prevalence of multiple sclerosis in Europe: A systematic review. BMC Neurol. 2013, 13, 128. [Google Scholar] [CrossRef] [PubMed]
- Kasper, L.H.; Shoemaker, J. Multiple sclerosis immunology: The healthy immune system vs. the MS immune system. Neurology 2010, 74 (Suppl. 1), S2–S8. [Google Scholar] [CrossRef] [PubMed]
- Katz Sand, I. Classification, diagnosis, and differential diagnosis of multiple sclerosis. Curr. Opin. Neurol. 2015, 28, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Housley, W.J.; Pitt, D.; Hafler, D.A. Biomarkers in multiple sclerosis. Clin. Immunol. 2015, 161, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Stangel, M.; Fredrikson, S.; Meinl, E.; Petzold, A.; Stuve, O.; Tumani, H. The utility of cerebrospinal fluid analysis in patients with multiple sclerosis. Nat. Rev. Neurol. 2013, 9, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Fisniku, L.K.; Brex, P.A.; Altmann, D.R.; Miszkiel, K.A.; Benton, C.E.; Lanyon, R.; Thompson, A.J.; Miller, D.H. Disability and T2 MRI lesions: A 20-year follow-up of patients with relapse onset of multiple sclerosis. Brain 2008, 131, 808–817. [Google Scholar] [CrossRef] [PubMed]
- Zivadinov, R.; Leist, T.P. Clinical-magnetic resonance imaging correlations in multiple sclerosis. J. Neuroimaging 2005, 15, 10S–21S. [Google Scholar] [CrossRef] [PubMed]
- Antoniol, C.; Stankoff, B. Immunological Markers for PML Prediction in MS Patients Treated with Natalizumab. Front. Immunol. 2014, 5, 668. [Google Scholar] [CrossRef] [PubMed]
- Outteryck, O.; Zephir, H.; Salleron, J.; Ongagna, J.C.; Etxeberria, A.; Collongues, N.; Lacour, A.; Fleury, M.C.; Blanc, F.; Giroux, M.; et al. JC-virus seroconversion in multiple sclerosis patients receiving natalizumab. Mult. Scler. 2014, 20, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Li, D.K.; Held, U.; Petkau, J.; Daumer, M.; Barkhof, F.; Fazekas, F.; Frank, J.A.; Kappos, L.; Miller, D.H.; Simon, J.H.; et al. MRI T2 lesion burden in multiple sclerosis: A plateauing relationship with clinical disability. Neurology 2006, 66, 1384–1389. [Google Scholar] [CrossRef]
- Plavina, T.; Subramanyam, M.; Bloomgren, G.; Richman, S.; Pace, A.; Lee, S.; Schlain, B.; Campagnolo, D.; Belachew, S.; Ticho, B. Anti-JC virus antibody levels in serum or plasma further define risk of natalizumab-associated progressive multifocal leukoencephalopathy. Ann. Neurol. 2014, 76, 802–812. [Google Scholar] [CrossRef] [Green Version]
- Rocca, M.A.; Amato, M.P.; de Stefano, N.; Enzinger, C.; Geurts, J.J.; Penner, I.K.; Rovira, A.; Sumowski, J.F.; Valsasina, P.; Filippi, M.; et al. Clinical and imaging assessment of cognitive dysfunction in multiple sclerosis. Lancet Neurol. 2015, 14, 302–317. [Google Scholar] [CrossRef]
- Sun, W.; YLi, S.J.; Huang, H.D.; Shyy, J.Y.; Chien, S. microRNA: A master regulator of cellular processes for bioengineering systems. Annu. Rev. Biomed. Eng. 2010, 12, 1–27. [Google Scholar] [CrossRef]
- Vannini, I.; Fanini, F.; Fabbri, M. Emerging roles of microRNAs in cancer. Curr. Opin. Genet. Dev. 2018, 48, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Pillai, R.S. MicroRNA function: Multiple mechanisms for a tiny RNA? RNA 2005, 11, 1753–1761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, A.; Baltimore, D. MicroRNAs as regulatory elements in immune system logic. Nat. Rev. Immunol. 2016, 16, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, M.; Croce, C.M. microRNAs: Master regulators as potential therapeutics in cancer. Annu. Rev. Pharmacol. Toxicol. 2011, 51, 25–43. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lan, H.Y.; Roukos, D.H.; Cho, W.C. Application of microRNAs in diabetes mellitus. J. Endocrinol. 2014, 222, R1–R10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrovic, N.; Ergun, S. miRNAs as Potential Treatment Targets and Treatment Options in Cancer. Mol. Diagn. Ther. 2018, 22, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Fenoglio, C.; Ridolfi, E.; Galimberti, D.; Scarpini, E. MicroRNAs as active players in the pathogenesis of multiple sclerosis. Int. J. Mol. Sci. 2012, 13, 13227–13239. [Google Scholar] [CrossRef] [PubMed]
- Vistbakka, J.; Elovaara, I.; Lehtimaki, T.; Hagman, S. Circulating microRNAs as biomarkers in progressive multiple sclerosis. Mult. Scler. 2017, 23, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Sode, J.; Krintel, S.B.; Carlsen, A.L.; Hetland, M.L.; Johansen, J.S.; Horslev-Petersen, K.; Stengaard-Pedersen, K.; Ellingsen, T.; Burton, M.; Junker, P.; et al. Plasma MicroRNA Profiles in Patients with Early Rheumatoid Arthritis Responding to Adalimumab plus Methotrexate vs Methotrexate Alone: A Placebo-controlled Clinical Trial. J. Rheumatol. 2018, 45, 53–61. [Google Scholar] [CrossRef]
- Stanczyk, J.; Pedrioli, D.M.; Brentano, F.; Sanchez-Pernaute, O.; Kolling, C.; Gay, R.E.; Detmar, M.; Gay, S.; Kyburz, D. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008, 58, 1001–1009. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Peng, W.; Ouyang, X.; Li, W.; Dai, Y. Circulating microRNAs as candidate biomarkers in patients with systemic lupus erythematosus. Transl. Res. 2012, 160, 198–206. [Google Scholar] [CrossRef]
- Dudics, S.; Venkatesha, S.H.; Moudgil, K.D. The Micro-RNA Expression Profiles of Autoimmune Arthritis Reveal Novel Biomarkers of the Disease and Therapeutic Response. Int. J. Mol. Sci. 2018, 19, 2293. [Google Scholar] [CrossRef] [PubMed]
- Constantinescu, C.S.; Farooqi, N.; O’Brien, K.; Gran, B. Experimental autoimmune encephalomyelitis (EAE) as a model for multiple sclerosis (MS). Br. J. Pharmacol. 2011, 164, 1079–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farooqi, N.; Gran, B.; Constantinescu, C.S. Are current disease-modifying therapeutics in multiple sclerosis justified on the basis of studies in experimental autoimmune encephalomyelitis? J. Neurochem. 2010, 115, 829–844. [Google Scholar] [CrossRef] [PubMed]
- Libbey, J.E.; Fujinami, R.S. Experimental autoimmune encephalomyelitis as a testing paradigm for adjuvants and vaccines. Vaccine 2011, 29, 3356–3362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linker, R.A.; Lee, D.H.; Demir, S.; Wiese, S.; Kruse, N.; Siglienti, I.; Gerhardt, E.; Neumann, H.; Sendtner, M.; Luhder, F.; et al. Functional role of brain-derived neurotrophic factor in neuroprotective autoimmunity: Therapeutic implications in a model of multiple sclerosis. Brain 2010, 133, 2248–2263. [Google Scholar] [CrossRef] [PubMed]
- Makar, T.K.; Nimmagadda, V.K.; Trisler, D.; Bever, C.T., Jr. Cell-based delivery of brain-derived neurotrophic factor in experimental allergic encephalomyelitis. J. Interferon Cytokine Res. 2014, 34, 641–647. [Google Scholar] [CrossRef] [PubMed]
- Butzkueven, H.; Emery, B.; Cipriani, T.; Marriott, M.P.; Kilpatrick, T.J. Endogenous leukemia inhibitory factor production limits autoimmune demyelination and oligodendrocyte loss. Glia 2006, 53, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Slaets, H.; Hendriks, J.J.; Stinissen, P.; Kilpatrick, T.J.; Hellings, N. Therapeutic potential of LIF in multiple sclerosis. Trends Mol. Med. 2010, 16, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Rot, U.; Sandelius, A.; Emersic, A.; Zetterberg, H.; Blennow, K. Cerebrospinal fluid GAP-43 in early multiple sclerosis. Mult. Scler. J. 2018, 4, 2055217318792931. [Google Scholar] [CrossRef] [PubMed]
- Lewczuk, P.; Ermann, N.; Andreasson, U.; Schultheis, C.; Podhorna, J.; Spitzer, P.; Maler, J.M.; Kornhuber, J.; Blennow, K.; Zetterberg, H. Plasma neurofilament light as a potential biomarker of neurodegeneration in Alzheimer’s disease. Alzheimer’s Res. Ther. 2018, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Lecca, D.; Marangon, D.; Coppolino, G.T.; Mendez, A.M.; Finardi, A.; Costa, G.D.; Martinelli, V.; Furlan, R.; Abbracchio, M.P. MiR-125a-3p timely inhibits oligodendroglial maturation and is pathologically up-regulated in human multiple sclerosis. Sci. Rep. 2016, 6, 34503. [Google Scholar] [CrossRef] [PubMed]
- Liguori, M.; Nuzziello, N.; Licciulli, F.; Consiglio, A.; Simone, M.; Viterbo, R.G.; Creanza, T.M.; Ancona, N.; Tortorella, C.; Margari, L.; et al. Combined microRNA and mRNA expression analysis in pediatric multiple sclerosis: An integrated approach to uncover novel pathogenic mechanisms of the disease. Hum. Mol. Genet. 2018, 27, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Reijerkerk, A.; Lopez-Ramirez, M.A.; van Het Hof, B.; Drexhage, J.A.; Kamphuis, W.W.; Kooij, G.; Vos, J.B.; van der Pouw Kraan, T.C.; van Zonneveld, A.J.; Horrevoets, A.J.; et al. MicroRNAs regulate human brain endothelial cell-barrier function in inflammation: Implications for multiple sclerosis. J. Neurosci. 2013, 33, 6857–6863. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Zhu, S.; Dai, D.; Liu, Z.; Li, D.; Li, B.; Gagliani, N.; Zheng, Y.; Tang, Y.; Weirauch, M.T.; et al. MiR-125a targets effector programs to stabilize Treg-mediated immune homeostasis. Nat. Commun. 2015, 6, 7096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, F.; Liu, T.; Sun, S.; Feng, S. The role of the miR-99b-5p/mTOR signaling pathway in neuroregeneration in mice following spinal cord injury. Mol. Med. Rep. 2017, 16, 9355–9360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, T.F.; Lin, J.F.; Chou, K.Y.; Lin, Y.C.; Chen, H.E.; Hwang, T.I. miR-99a-5p acts as tumor suppressor via targeting to mTOR and enhances RAD001-induced apoptosis in human urinary bladder urothelial carcinoma cells. Onco Targets Ther. 2018, 11, 239–252. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Kong, C.; Tsun, A.; Chen, C.; Song, H.; Shi, G.; Pan, W.; Dai, D.; Shen, N.; Li, B. MiR-125a-5p Decreases the Sensitivity of Treg cells Toward IL-6-Mediated Conversion by Inhibiting IL-6R and STAT3 Expression. Sci. Rep. 2015, 5, 14615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Cheng, Y.; Cui, W.; Li, M.; Li, B.; Guo, L. MicroRNA-155 modulates Th1 and Th17 cell differentiation and is associated with multiple sclerosis and experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2014, 266, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Murugaiyan, G.; Beynon, V.; Mittal, A.; Joller, N.; Weiner, H.L. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J. Immunol. 2011, 187, 2213–2221. [Google Scholar] [CrossRef]
- Ksiazek-Winiarek, D.; Szpakowski, P.; Turniak, M.; Szemraj, J.; Glabinski, A. IL-17 Exerts Anti-Apoptotic Effect via miR-155-5p Downregulation in Experimental Autoimmune Encephalomyelitis. J. Mol. Neurosci. 2017, 63, 320–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Ramirez, M.A.; Wu, D.; Pryce, G.; Simpson, J.E.; Reijerkerk, A.; King-Robson, J.; Kay, O.; de Vries, H.E.; Hirst, M.C.; Sharrack, B.; et al. MicroRNA-155 negatively affects blood-brain barrier function during neuroinflammation. FASEB J. 2014, 28, 2551–2565. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhou, J.; Zhong, Y.; Jiang, L.; Mu, P.; Li, Y.; Singh, N.; Nagarkatti, M.; Nagarkatti, P. Expression, regulation and function of microRNAs in multiple sclerosis. Int. J. Med. Sci. 2014, 11, 810–818. [Google Scholar] [CrossRef] [PubMed]
- Baulina, N.; Kulakova, O.; Kiselev, I.; Osmak, G.; Popova, E.; Boyko, A.; Favorova, O. Immune-related miRNA expression patterns in peripheral blood mononuclear cells differ in multiple sclerosis relapse and remission. J. Neuroimmunol. 2018, 317, 67–76. [Google Scholar] [CrossRef] [PubMed]
- McCoy, C.E. miR-155 Dysregulation and Therapeutic Intervention in Multiple Sclerosis. Adv. Exp. Med. Biol. 2017, 1024, 111–131. [Google Scholar] [PubMed]
- Guan, H.; Fan, D.; Mrelashvili, D.; Hao, H.; Singh, N.P.; Singh, U.P.; Nagarkatti, P.S.; Nagarkatti, M. MicroRNA let-7e is associated with the pathogenesis of experimental autoimmune encephalomyelitis. Eur. J. Immunol. 2013, 43, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Han, J.J.; Liang, X.Y.; Zhao, L.; Zhang, F.; Rasouli, J.; Wang, Z.Z.; Zhang, G.X.; Li, X. miR-23b Suppresses Leukocyte Migration and Pathogenesis of Experimental Autoimmune Encephalomyelitis by Targeting CCL7. Mol. Ther. 2018, 26, 582–592. [Google Scholar] [CrossRef]
- Hoye, M.L.; Archambault, A.S.; Gordon, T.M.; Oetjen, L.K.; Cain, M.D.; Klein, R.S.; Crosby, S.D.; Kim, B.S.; Miller, T.M.; Wu, G.F. MicroRNA signature of central nervous system-infiltrating dendritic cells in an animal model of multiple sclerosis. Immunology 2018. [Google Scholar] [CrossRef]
- Zhu, S.; Pan, W.; Song, X.; Liu, Y.; Shao, X.; Tang, Y.; Liang, D.; He, D.; Wang, H.; Liu, W.; et al. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-alpha. Nat. Med. 2012, 18, 1077–1086. [Google Scholar] [CrossRef]
- Bhairavabhotla, R.; Kim, Y.C.; Glass, D.D.; Escobar, T.M.; Patel, M.C.; Zahr, R.; Nguyen, C.K.; Kilaru, G.K.; Muljo, S.A.; Shevach, E.M. Transcriptome profiling of human FoxP3+ regulatory T cells. Hum. Immunol. 2016, 77, 201–213. [Google Scholar] [CrossRef] [Green Version]
- Ifergan, I.; Chen, S.; Zhang, B.; Miller, S.D. Cutting Edge: MicroRNA-223 Regulates Myeloid Dendritic Cell-Driven Th17 Responses in Experimental Autoimmune Encephalomyelitis. J. Immunol. 2009, 196, 1455–1459. [Google Scholar] [CrossRef]
- Chen, J.; Adamiak, W.; Huang, G.; Atasoy, U.; Rostami, A.; Yu, S. Interaction of RNA-binding protein HuR and miR-466i regulates GM-CSF expression. Sci. Rep. 2017, 7, 17233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarthy, D.P.; Richards, M.H.; Miller, S.D. Mouse models of multiple sclerosis: Experimental autoimmune encephalomyelitis and Theiler’s virus-induced demyelinating disease. Methods Mol. Biol. 2012, 900, 381–401. [Google Scholar] [PubMed]
- Li, X.; Lees, J.R. Pre-existing central nervous system lesions negate cytokine requirements for regional experimental autoimmune encephalomyelitis development. Immunology 2013, 138, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Tackett, M.R.; Diwan, I. Using FirePlex() Particle Technology for Multiplex MicroRNA Profiling Without RNA Purification. Methods Mol. Biol. 2017, 1654, 209–219. [Google Scholar] [PubMed]

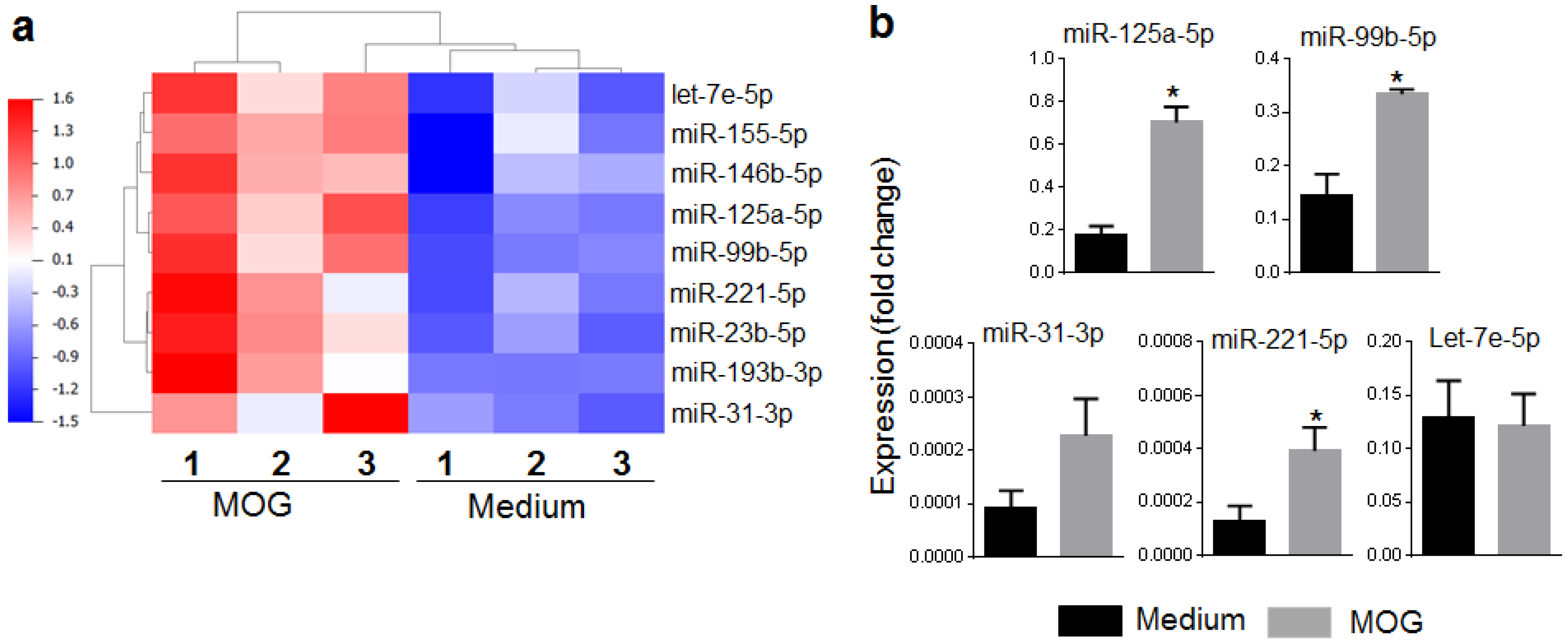





| miRNA | mRNA Targets |
|---|---|
| Let-7e-5p | AGO2, BCL2L1, BSG, CCL3, CCR7, CD200, CD28, CD80, CD86, CHUK, CRP, DUSP1, EPHA4, FASLG, IL12RB2, IL13, IL6, ITGB8, LYN, MAPK11, PRDM1, RORC, TLR4, TNFRSF1B, TNFSF10 |
| miR-155-5p | CTLA4, CD274, CD47, HIF1A, RELA, SMAD4, ETS1, NDFIP1, IL21, ITK, S1PR1, BDNF |
| miR-146b-5p | AGO2, CCR3, CCR5, CD40, CHUCK, CRP, CXCL8, CXCR4, GDNF, IFNA1/IFNA13, IFNB1, IKZF4, IL10, IL17A, IL1R1, IL2RB2, IRAK1, IRAK2, IRAK2, MAPK14, NOS2, PLP1, SP1, STAT1, STAT3, TLR1, TLR10, TLR4, TLR9, TRAF6 |
| miR-125a-5p | AGO2, BDNF, CCR5, CSNK2A1, ELAVL1, ETS1, IKZFA, IL10RA, IL1RN, IL31, IL6R, IRAK1, IRF4, LIF, MAP2K7, MAP3K10, MAP3K11, MAPK12, MAPK14, MYD88, NOS2, PRDM1, RPS6KA1, SEMA4D, SGPL1, STAT3, TLR4, TNFRSF1B, TNFSF4, TP53, VTCN1 |
| miR-99b-5p | AGO2, FGFR3, MTOR, NTRK3, IGF1R, LIF, PRDM1, IKZF4 |
| miR-221-5p | IFNAR1, HTT, IL6ST, BDNF, IRF4 |
| miR-23b-5p | CCL7, FOXO4, IKKA, SMAD3, TAB2, TBA3 |
| miR-193b-3p | KIT, SOX5, BCL2L2, ETS1, SOCS3, ALOX5, KIT |
| miR-31-3p | PRDM1, NFATC2, HAVCR2, STAT3, FOXO1, FOXP3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venkatesha, S.H.; Dudics, S.; Song, Y.; Mahurkar, A.; Moudgil, K.D. The miRNA Expression Profile of Experimental Autoimmune Encephalomyelitis Reveals Novel Potential Disease Biomarkers. Int. J. Mol. Sci. 2018, 19, 3990. https://doi.org/10.3390/ijms19123990
Venkatesha SH, Dudics S, Song Y, Mahurkar A, Moudgil KD. The miRNA Expression Profile of Experimental Autoimmune Encephalomyelitis Reveals Novel Potential Disease Biomarkers. International Journal of Molecular Sciences. 2018; 19(12):3990. https://doi.org/10.3390/ijms19123990
Chicago/Turabian StyleVenkatesha, Shivaprasad H., Steven Dudics, Yang Song, Anup Mahurkar, and Kamal D. Moudgil. 2018. "The miRNA Expression Profile of Experimental Autoimmune Encephalomyelitis Reveals Novel Potential Disease Biomarkers" International Journal of Molecular Sciences 19, no. 12: 3990. https://doi.org/10.3390/ijms19123990





