Decreased PEDF Expression Promotes Adipogenic Differentiation through the Up-Regulation of CD36
Abstract
:1. Introduction
2. Results
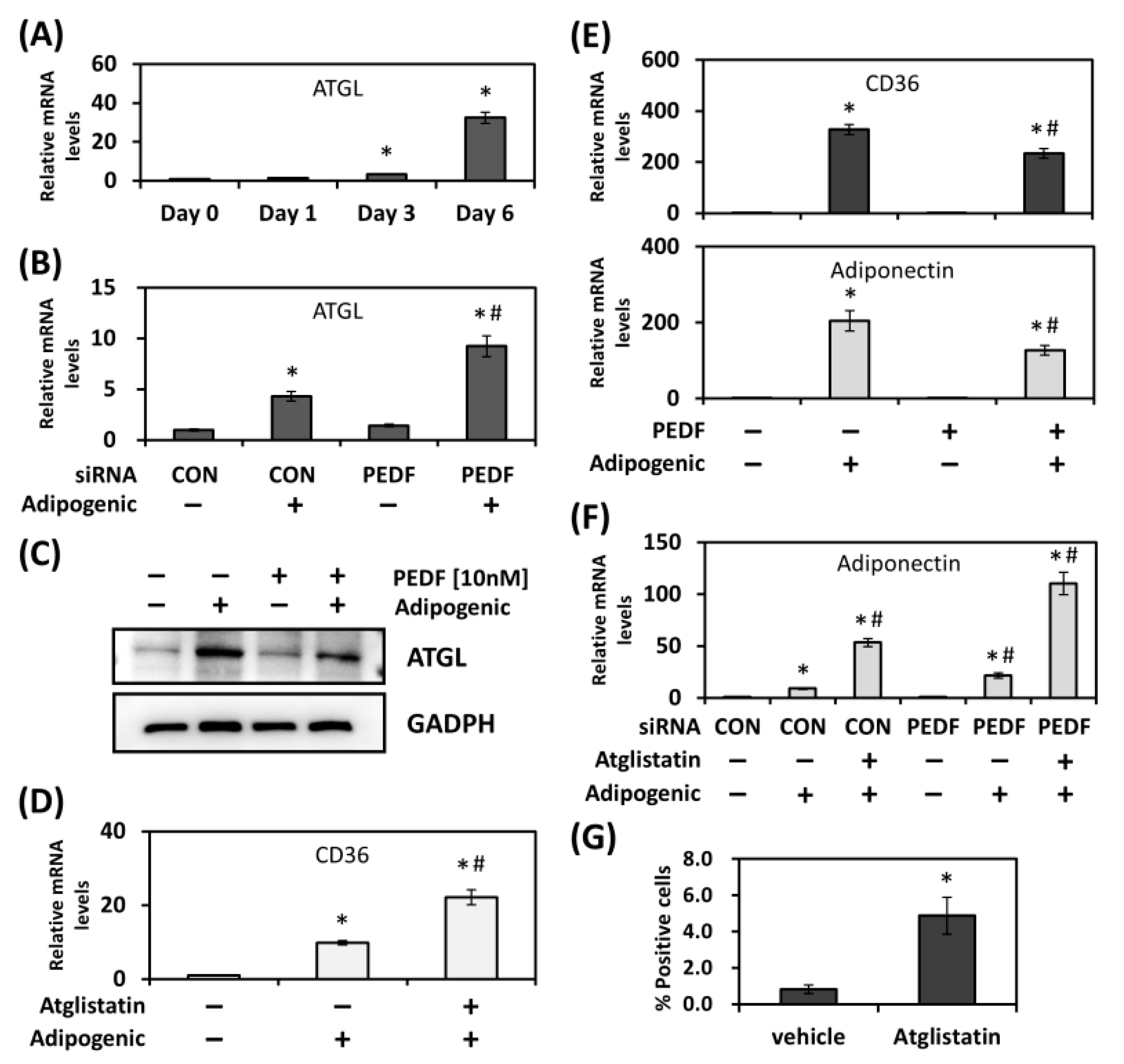
2.1. PEDF Regulates CD36 Expression during Adipogenic Differentiation
2.2. Inhibition of ATGL Activity Promotes CD36 Expression and Adipogenesis
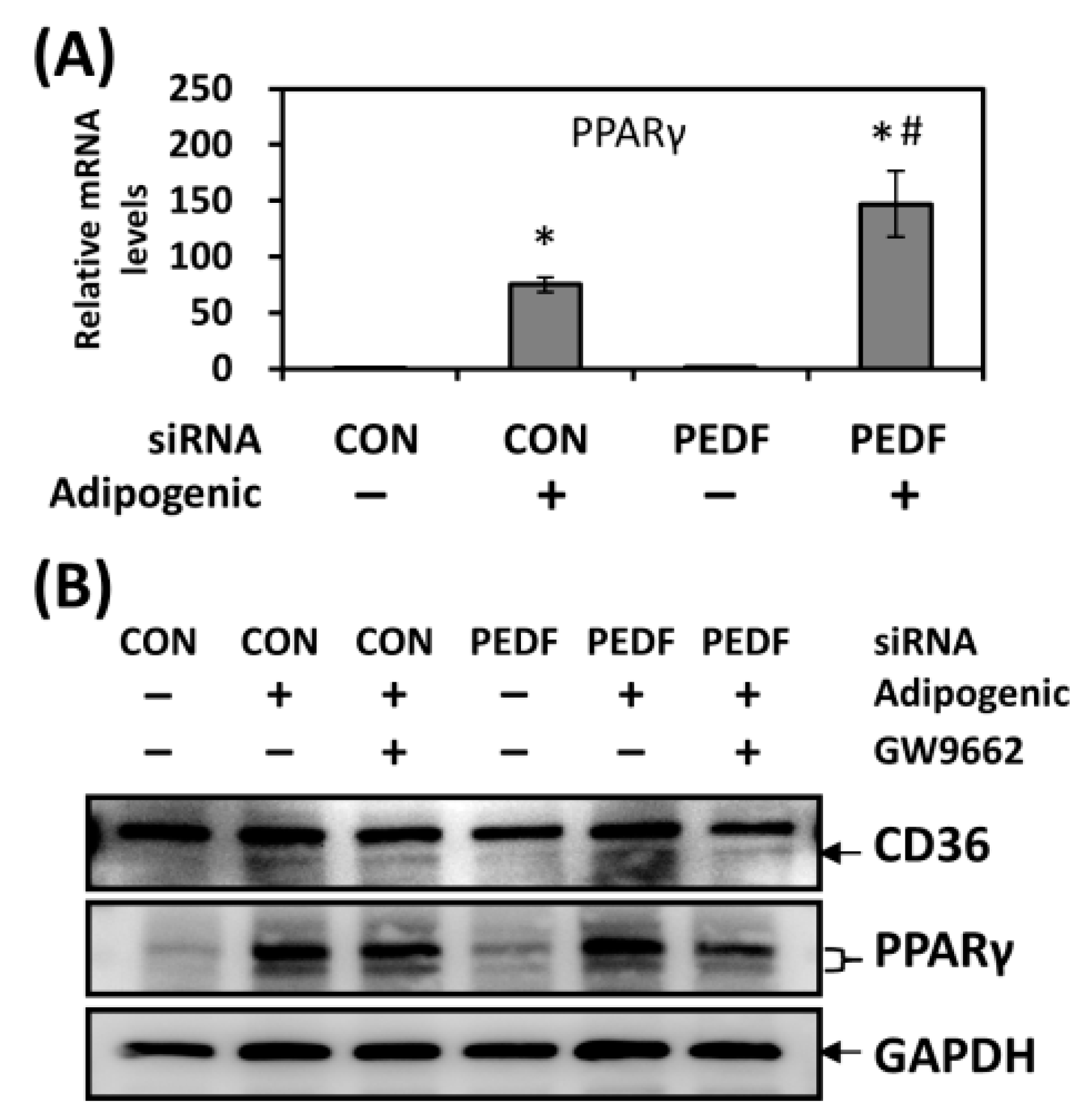
2.3. PEDF Decrease during Adipogenesis Leads to Increased Levels of PPARγ, a Positive Regulator of CD36
2.4. CD36 Regulates Mitotic Clonal Expansion and Lipogenic Genes during Adipogenic Differentiation
2.5. Glucocorticoids Regulate PEDF Transcription during Adipogenesis
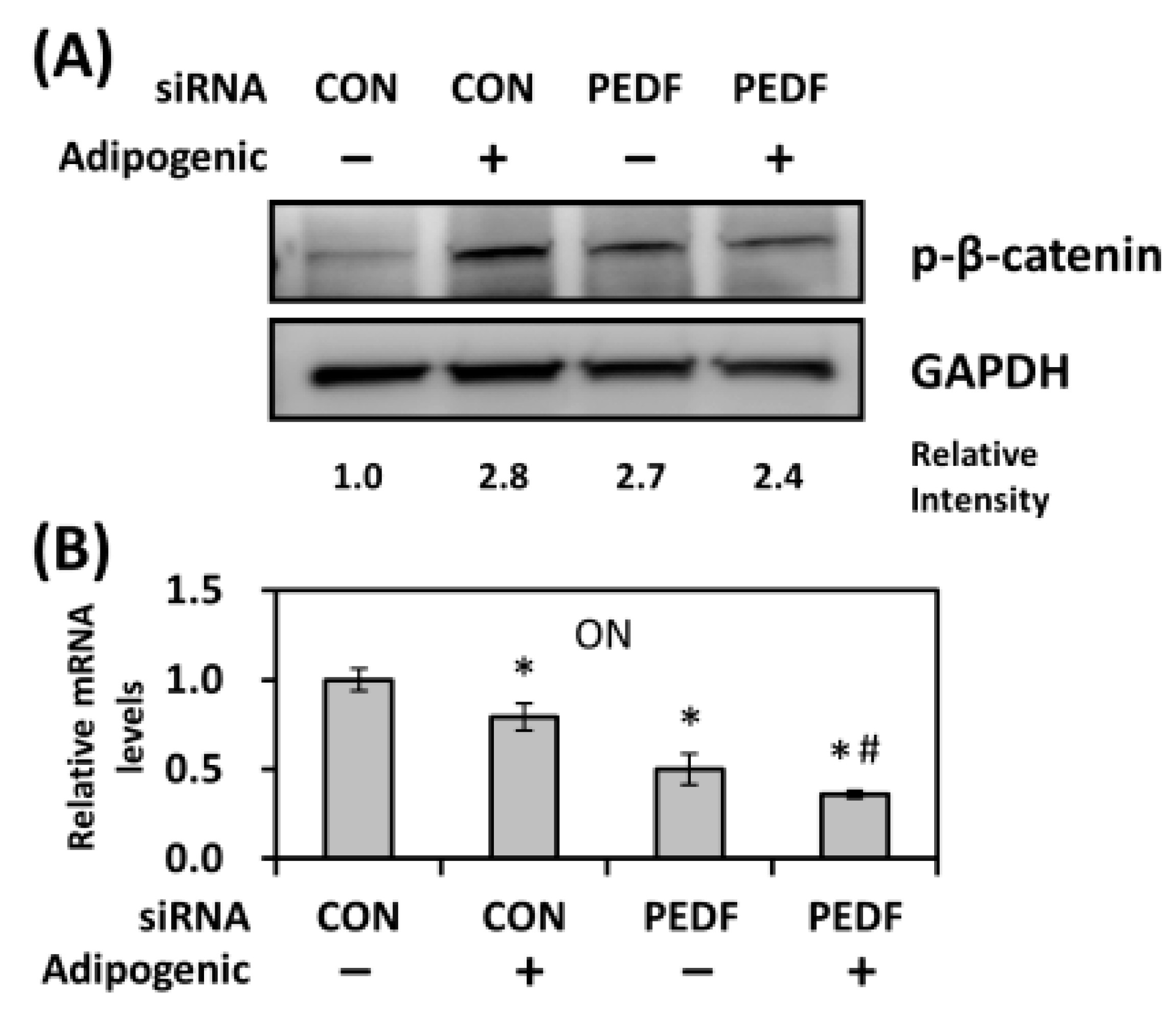
2.6. Decreased PEDF Causes an Increase in β-Catenin Phosphorylation during Adipogenesis
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Treatments
4.2. BODIPY 493/503 Staining
4.3. Quantitative RT-PCR
4.4. Immunoblotting
4.5. Cell Viability Assay
4.6. PEDF Promoter Assay
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PEDF | pigment epithelium-derived factor |
| ATGL | adipose triglyceride lipase |
| AdSCs | adipose-derived stem cells |
| CD36 | cluster of differentiation 36 |
| PPARγ | peroxisome proliferator-activated receptor gamma |
| C/EBP-α | CCAAT/enhancer-binding protein-alpha |
| SSO | sulfo-N-succinimidyl oleate |
| FASN | fatty acid synthase |
| SCD1 | stearoyl-CoA desaturase 1 |
| ACC1 | acetyl-CoA carboxylase 1 |
| NAFLD | non-alcoholic fatty liver disease |
| IBMX | 3-isobutyl-1-methylxanthine |
| ON | osteonectin |
References
- Rodriguez-Hernandez, H.; Simental-Mendia, L.E.; Rodriguez-Ramirez, G.; Reyes-Romero, M.A. Obesity and inflammation: Epidemiology, risk factors, and markers of inflammation. Int. J. Endocrinol. 2013, 2013, 678159. [Google Scholar] [CrossRef] [PubMed]
- Jo, J.; Gavrilova, O.; Pack, S.; Jou, W.; Mullen, S.; Sumner, A.E.; Cushman, S.W.; Periwal, V. Hypertrophy and/or Hyperplasia: Dynamics of Adipose Tissue Growth. PLoS Comput. Boil. 2009, 5, e1000324. [Google Scholar] [CrossRef] [PubMed]
- Drevon, C.A. Fatty acids and expression of adipokines. Biochim. Biophys. Acta 2005, 1740, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Giralt, M.; Cereijo, R.; Villarroya, F. Adipokines and the Endocrine Role of Adipose Tissues. Handb. Exp. Pharmacol. 2016, 233, 265–282. [Google Scholar] [CrossRef] [PubMed]
- Guilherme, A.; Virbasius, J.V.; Puri, V.; Czech, M.P. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nature reviews. Mol. Cell Boil. 2008, 9, 367–377. [Google Scholar] [CrossRef]
- Mancuso, P. The role of adipokines in chronic inflammation. ImmunoTargets Ther. 2016, 5, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Becerra, S.P.; Sagasti, A.; Spinella, P.; Notario, V. Pigment epithelium-derived factor behaves like a noninhibitory serpin. Neurotrophic activity does not require the serpin reactive loop. J. Boil. Chem. 1995, 270, 25992–25999. [Google Scholar] [CrossRef]
- Steele, F.R.; Chader, G.J.; Johnson, L.V.; Tombran-Tink, J. Pigment epithelium-derived factor: Neurotrophic activity and identification as a member of the serine protease inhibitor gene family. Proc. Natl. Acad. Sci. USA 1993, 90, 1526–1530. [Google Scholar] [CrossRef] [PubMed]
- Dawson, D.W.; Volpert, O.V.; Gillis, P.; Crawford, S.E.; Xu, H.; Benedict, W.; Bouck, N.P. Pigment epithelium-derived factor: A potent inhibitor of angiogenesis. Science 1999, 285, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Doll, J.A.; Gattu, A.K.; Shugrue, C.; Cornwell, M.; Fitchev, P.; Crawford, S.E. Anti-angiogenic pigment epithelium-derived factor regulates hepatocyte triglyceride content through adipose triglyceride lipase (ATGL). J. Hepatol. 2008, 48, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Gattu, A.K.; Swenson, E.S.; Iwakiri, Y.; Samuel, V.T.; Troiano, N.; Berry, R.; Church, C.D.; Rodeheffer, M.S.; Carpenter, T.O.; Chung, C. Determination of mesenchymal stem cell fate by pigment epithelium-derived factor (PEDF) results in increased adiposity and reduced bone mineral content. FASEB J. 2013, 27, 4384–4394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Wang, J.J.; Li, J.; Park, K.; Qian, X.; Ma, J.X.; Zhang, S.X. Pigment epithelium-derived factor suppresses adipogenesis via inhibition of the MAPK/ERK pathway in 3T3-L1 preadipocytes. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E1378–E1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crowe, S.; Wu, L.E.; Economou, C.; Turpin, S.M.; Matzaris, M.; Hoehn, K.L.; Hevener, A.L.; James, D.E.; Duh, E.J.; Watt, M.J. Pigment epithelium-derived factor contributes to insulin resistance in obesity. Cell Metab. 2009, 10, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, S.; Adachi, H.; Abe, A.; Yashiro, T.; Enomoto, M.; Furuki, K.; Hino, A.; Jinnouchi, Y.; Takenaka, K.; Matsui, T.; et al. Elevated serum levels of pigment epithelium-derived factor in the metabolic syndrome. J. Clin. Endocrinol. Metab. 2006, 91, 2447–2450. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.T.; Lin, C.C.; Tsai, M.C.; Chen, K.D.; Chiu, K.W. Pigment epithelium-derived factor in lipid metabolic disorders. Biomed. J. 2018, 41, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Famulla, S.; Lamers, D.; Hartwig, S.; Passlack, W.; Horrighs, A.; Cramer, A.; Lehr, S.; Sell, H.; Eckel, J. Pigment epithelium-derived factor (PEDF) is one of the most abundant proteins secreted by human adipocytes and induces insulin resistance and inflammatory signaling in muscle and fat cells. Int. J. Obes. 2011, 35, 762–772. [Google Scholar] [CrossRef] [PubMed]
- Gattu, A.K.; Birkenfeld, A.L.; Iwakiri, Y.; Jay, S.; Saltzman, M.; Doll, J.; Protiva, P.; Samuel, V.T.; Crawford, S.E.; Chung, C. Pigment epithelium-derived factor (PEDF) suppresses IL-1beta-mediated c-Jun N-terminal kinase (JNK) activation to improve hepatocyte insulin signaling. Endocrinology 2014, 155, 1373–1385. [Google Scholar] [CrossRef]
- Yoshida, T.; Yamagishi, S.; Nakamura, K.; Matsui, T.; Imaizumi, T.; Takeuchi, M.; Koga, H.; Ueno, T.; Sata, M. Pigment epithelium-derived factor (PEDF) ameliorates advanced glycation end product (AGE)-induced hepatic insulin resistance in vitro by suppressing Rac-1 activation. Horm. Metab. Res. 2008, 40, 620–625. [Google Scholar] [CrossRef]
- Zhou, J.; Febbraio, M.; Wada, T.; Zhai, Y.; Kuruba, R.; He, J.; Lee, J.H.; Khadem, S.; Ren, S.; Li, S.; et al. Hepatic fatty acid transporter Cd36 is a common target of LXR, PXR, and PPARgamma in promoting steatosis. Gastroenterology 2008, 134, 556–567. [Google Scholar] [CrossRef]
- Tang, Q.Q.; Otto, T.C.; Lane, M.D. Mitotic clonal expansion: A synchronous process required for adipogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 44–49. [Google Scholar] [CrossRef]
- Wiper-Bergeron, N.; Salem, H.A.; Tomlinson, J.J.; Wu, D.; Hache, R.J. Glucocorticoid-stimulated preadipocyte differentiation is mediated through acetylation of C/EBPbeta by GCN5. Proc. Natl. Acad. Sci. USA 2007, 104, 2703–2708. [Google Scholar] [CrossRef]
- Khan, A.; Fornes, O.; Stigliani, A.; Gheorghe, M.; Castro-Mondragon, J.A.; van der Lee, R.; Bessy, A.; Cheneby, J.; Kulkarni, S.R.; Tan, G.; et al. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018, 46, D260–D266. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.N.; Ross, S.E.; Longo, K.A.; Bajnok, L.; Hemati, N.; Johnson, K.W.; Harrison, S.D.; MacDougald, O.A. Regulation of Wnt signaling during adipogenesis. J. Boil. Chem. 2002, 277, 30998–31004. [Google Scholar] [CrossRef] [PubMed]
- Gaur, T.; Lengner, C.J.; Hovhannisyan, H.; Bhat, R.A.; Bodine, P.V.; Komm, B.S.; Javed, A.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J. Boil. Chem. 2005, 280, 33132–33140. [Google Scholar] [CrossRef] [PubMed]
- MacDougald, O.A.; Lane, M.D. Transcriptional regulation of gene expression during adipocyte differentiation. Annu. Rev. Biochem. 1995, 64, 345–373. [Google Scholar] [CrossRef]
- Haemmerle, G.; Zimmermann, R.; Strauss, J.G.; Kratky, D.; Riederer, M.; Knipping, G.; Zechner, R. Hormone-sensitive lipase deficiency in mice changes the plasma lipid profile by affecting the tissue-specific expression pattern of lipoprotein lipase in adipose tissue and muscle. J. Boil. Chem. 2002, 277, 12946–12952. [Google Scholar] [CrossRef]
- Notari, L.; Baladron, V.; Aroca-Aguilar, J.D.; Balko, N.; Heredia, R.; Meyer, C.; Notario, P.M.; Saravanamuthu, S.; Nueda, M.L.; Sanchez-Sanchez, F.; et al. Identification of a lipase-linked cell membrane receptor for pigment epithelium-derived factor. J. Boil. Chem. 2006, 281, 38022–38037. [Google Scholar] [CrossRef]
- Haemmerle, G.; Lass, A.; Zimmermann, R.; Gorkiewicz, G.; Meyer, C.; Rozman, J.; Heldmaier, G.; Maier, R.; Theussl, C.; Eder, S.; et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 2006, 312, 734–737. [Google Scholar] [CrossRef]
- Dai, Z.; Qi, W.; Li, C.; Lu, J.; Mao, Y.; Yao, Y.; Li, L.; Zhang, T.; Hong, H.; Li, S.; et al. Dual regulation of adipose triglyceride lipase by pigment epithelium-derived factor: A novel mechanistic insight into progressive obesity. Mol. Cell. Endocrinol. 2013, 377, 123–134. [Google Scholar] [CrossRef]
- Kim, J.Y.; Tillison, K.; Lee, J.H.; Rearick, D.A.; Smas, C.M. The adipose tissue triglyceride lipase ATGL/PNPLA2 is downregulated by insulin and TNF-alpha in 3T3-L1 adipocytes and is a target for transactivation by PPARgamma. American journal of physiology. Endocrinol. Metab. 2006, 291, E115–E127. [Google Scholar] [CrossRef]
- Zechner, R.; Zimmermann, R.; Eichmann, T.O.; Kohlwein, S.D.; Haemmerle, G.; Lass, A.; Madeo, F. FAT SIGNALS—Lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012, 15, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Febbraio, M.; Hajjar, D.P.; Silverstein, R.L. CD36: A class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J. Clin. Investing. 2001, 108, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Melnichenko, A.A.; Myasoedova, V.A.; Grechko, A.V.; Orekhov, A.N. Mechanisms of foam cell formation in atherosclerosis. J. Mol. Med. 2017, 95, 1153–1165. [Google Scholar] [CrossRef] [PubMed]
- Hosui, A.; Tatsumi, T.; Hikita, H.; Saito, Y.; Hiramatsu, N.; Tsujii, M.; Hennighausen, L.; Takehara, T. Signal transducer and activator of transcription 5 plays a crucial role in hepatic lipid metabolism through regulation of CD36 expression. Hepatol. Res. 2016, 47, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Koonen, D.P.; Jacobs, R.L.; Febbraio, M.; Young, M.E.; Soltys, C.L.; Ong, H.; Vance, D.E.; Dyck, J.R. Increased hepatic CD36 expression contributes to dyslipidemia associated with diet-induced obesity. Diabetes 2007, 56, 2863–2871. [Google Scholar] [CrossRef] [PubMed]
- Sheedfar, F.; Sung, M.M.; Aparicio-Vergara, M.; Kloosterhuis, N.J.; Miquilena-Colina, M.E.; Vargas-Castrillon, J.; Febbraio, M.; Jacobs, R.L.; de Bruin, A.; Vinciguerra, M.; et al. Increased hepatic CD36 expression with age is associated with enhanced susceptibility to nonalcoholic fatty liver disease. Aging 2014, 6, 281–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christiaens, V.; Van Hul, M.; Lijnen, H.R.; Scroyen, I. CD36 promotes adipocyte differentiation and adipogenesis. Biochim. Biophys. Acta 2012, 1820, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Collins, J.M.; Neville, M.J.; Pinnick, K.E.; Hodson, L.; Ruyter, B.; van Dijk, T.H.; Reijngoud, D.J.; Fielding, M.D.; Frayn, K.N. De novo lipogenesis in the differentiating human adipocyte can provide all fatty acids necessary for maturation. J. Lipid Res. 2011, 52, 1683–1692. [Google Scholar] [CrossRef] [Green Version]
- Johannsson, G.; Ragnarsson, O. Cardiovascular and metabolic impact of glucocorticoid replacement therapy. Front. Horm. Res. 2014, 43, 33–44. [Google Scholar] [CrossRef]
- Chapman, A.B.; Knight, D.M.; Ringold, G.M. Glucocorticoid regulation of adipocyte differentiation: Hormonal triggering of the developmental program and induction of a differentiation-dependent gene. J. Cell Boil. 1985, 101, 1227–1235. [Google Scholar] [CrossRef]
- Daubriac, J.; Pandya, U.M.; Huang, K.T.; Pavlides, S.C.; Gama, P.; Blank, S.V.; Shukla, P.; Crawford, S.E.; Gold, L.I. Hormonal and Growth Regulation of Epithelial and Stromal Cells From the Normal and Malignant Endometrium by Pigment Epithelium-Derived Factor. Endocrinology 2017, 158, 2754–2773. [Google Scholar] [CrossRef] [PubMed]
- Doll, J.A.; Stellmach, V.M.; Bouck, N.P.; Bergh, A.R.; Lee, C.; Abramson, L.P.; Cornwell, M.L.; Pins, M.R.; Borensztajn, J.; Crawford, S.E. Pigment epithelium-derived factor regulates the vasculature and mass of the prostate and pancreas. Nat. Med. 2003, 9, 774–780. [Google Scholar] [CrossRef] [PubMed]
- Tombran-Tink, J.; Lara, N.; Apricio, S.E.; Potluri, P.; Gee, S.; Ma, J.X.; Chader, G.; Barnstable, C.J. Retinoic acid and dexamethasone regulate the expression of PEDF in retinal and endothelial cells. Exp. Eye Res. 2004, 78, 945–955. [Google Scholar] [CrossRef] [PubMed]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, K.-T.; Hsu, L.-W.; Chen, K.-D.; Kung, C.-P.; Goto, S.; Chen, C.-L. Decreased PEDF Expression Promotes Adipogenic Differentiation through the Up-Regulation of CD36. Int. J. Mol. Sci. 2018, 19, 3992. https://doi.org/10.3390/ijms19123992
Huang K-T, Hsu L-W, Chen K-D, Kung C-P, Goto S, Chen C-L. Decreased PEDF Expression Promotes Adipogenic Differentiation through the Up-Regulation of CD36. International Journal of Molecular Sciences. 2018; 19(12):3992. https://doi.org/10.3390/ijms19123992
Chicago/Turabian StyleHuang, Kuang-Tzu, Li-Wen Hsu, Kuang-Den Chen, Chao-Pin Kung, Shigeru Goto, and Chao-Long Chen. 2018. "Decreased PEDF Expression Promotes Adipogenic Differentiation through the Up-Regulation of CD36" International Journal of Molecular Sciences 19, no. 12: 3992. https://doi.org/10.3390/ijms19123992




