OsGPAT3 Plays a Critical Role in Anther Wall Programmed Cell Death and Pollen Development in Rice
Abstract
:1. Introduction
2. Results
2.1. Isolation and Phenotypic Analyses of the gpat3-2 Mutant
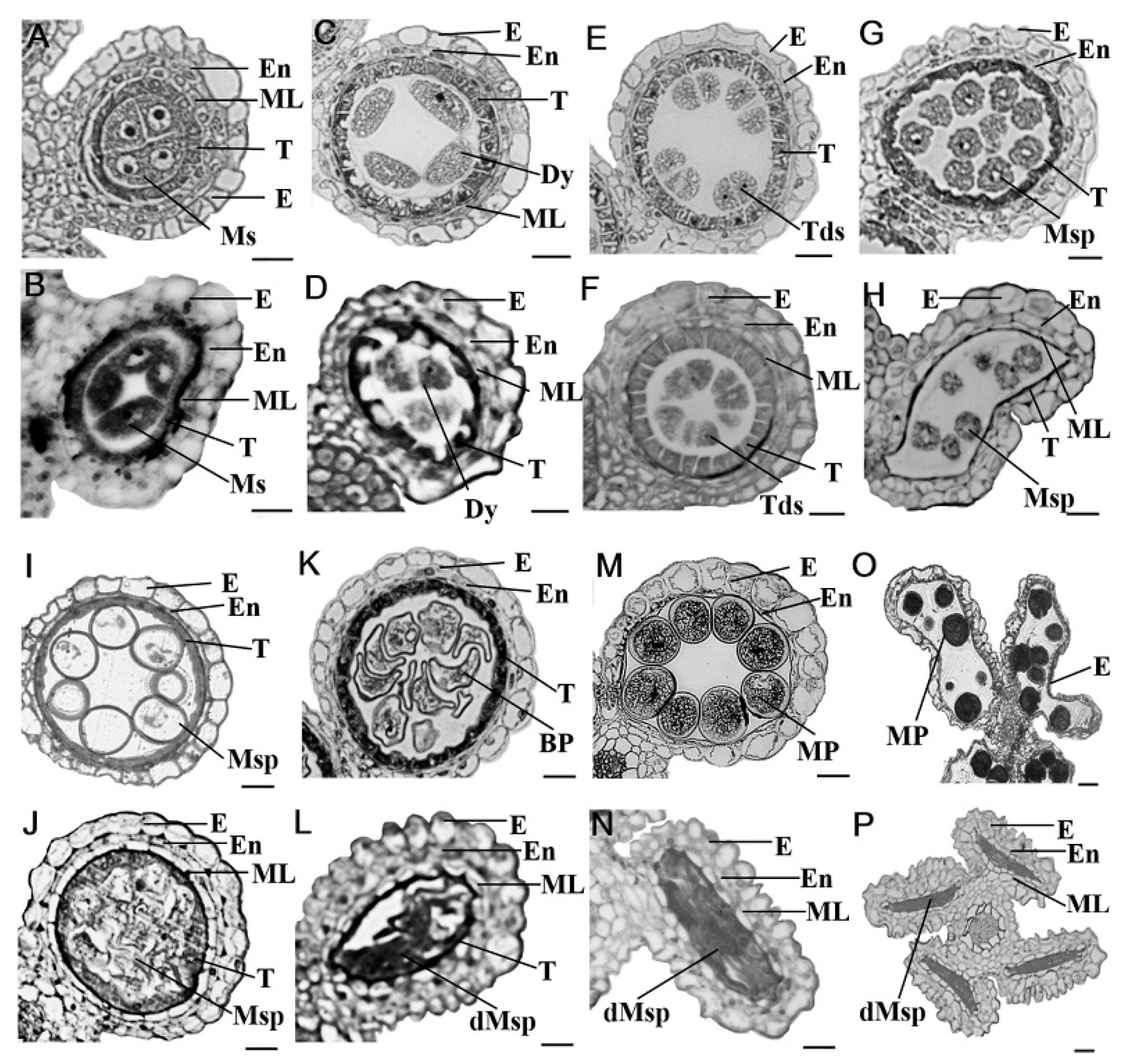
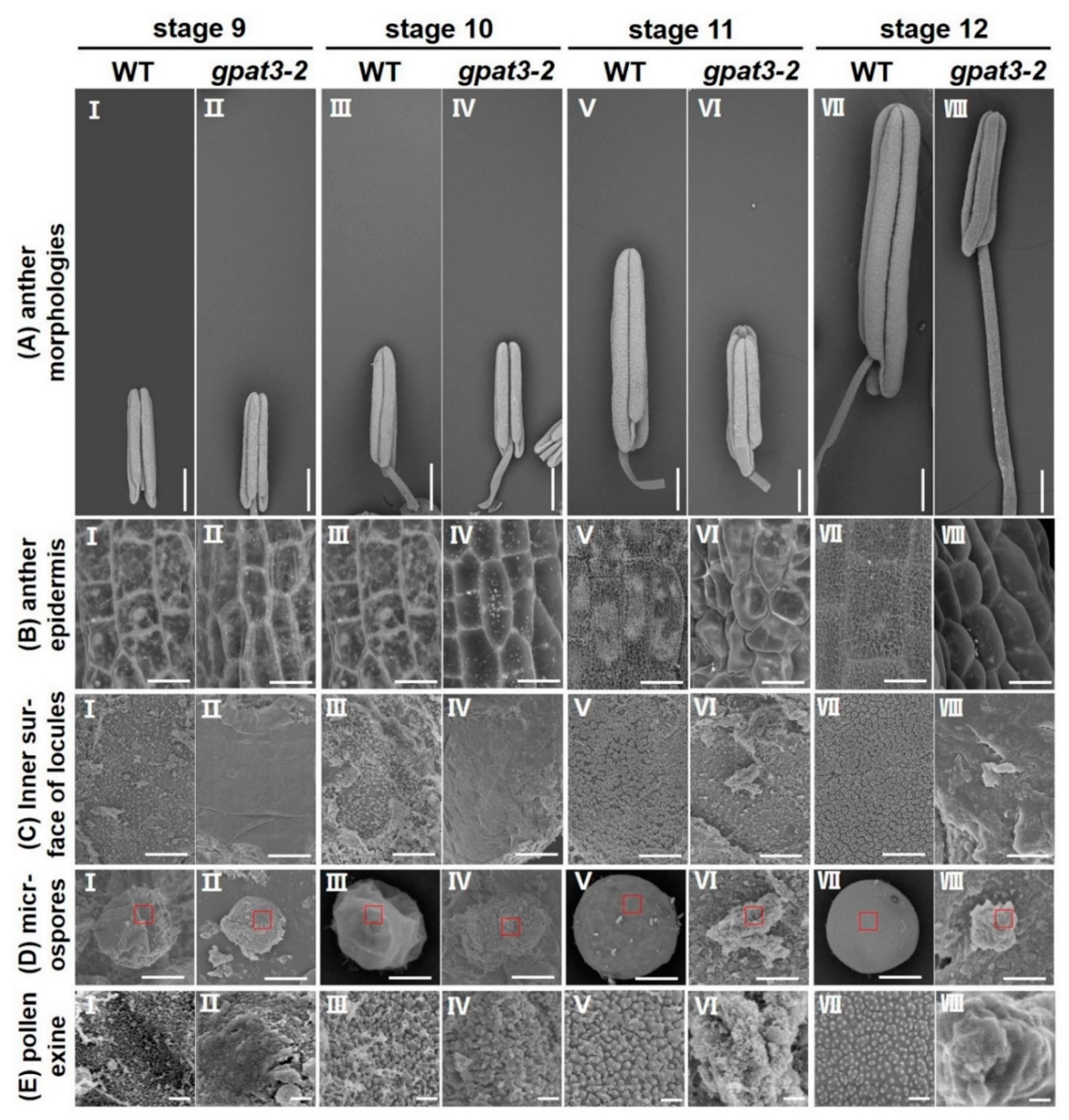
2.2. Defects of Anther Development and Pollen Maturation in gpat3-2 Mutant
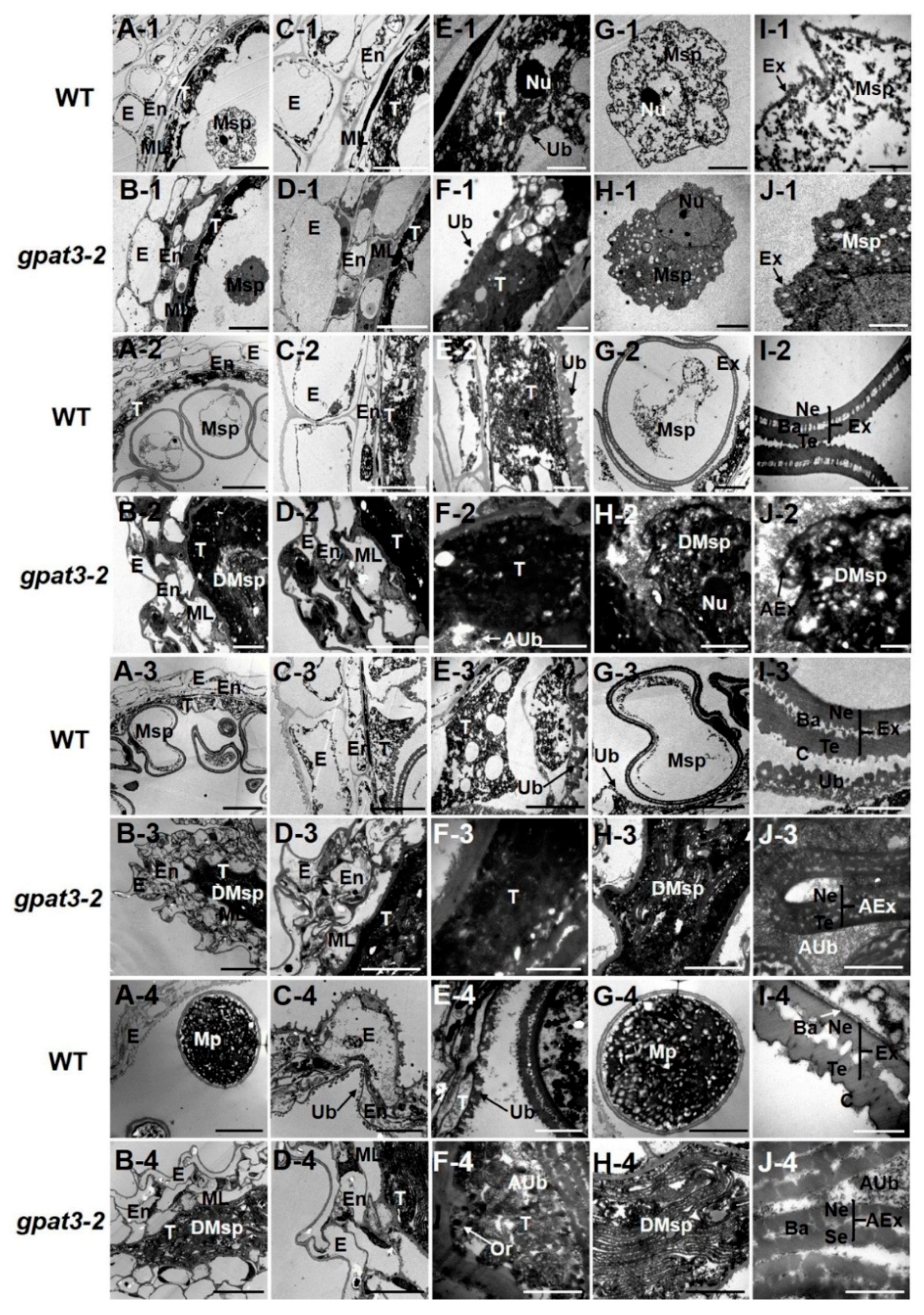
2.3. Delayed PCD of Osgpat3-2 Tapetal Cells and Anther Wall Cells
2.4. Osgpat3-2 Exhibits Defects in the Formation of Ubisch Bodies and Pollen Accumulation
2.5. Fine Genetic Mapping and Candidate Gene Analysis of the Osgpat3-2 Mutation
2.6. Function Verification of the OsGPAT3 Gene
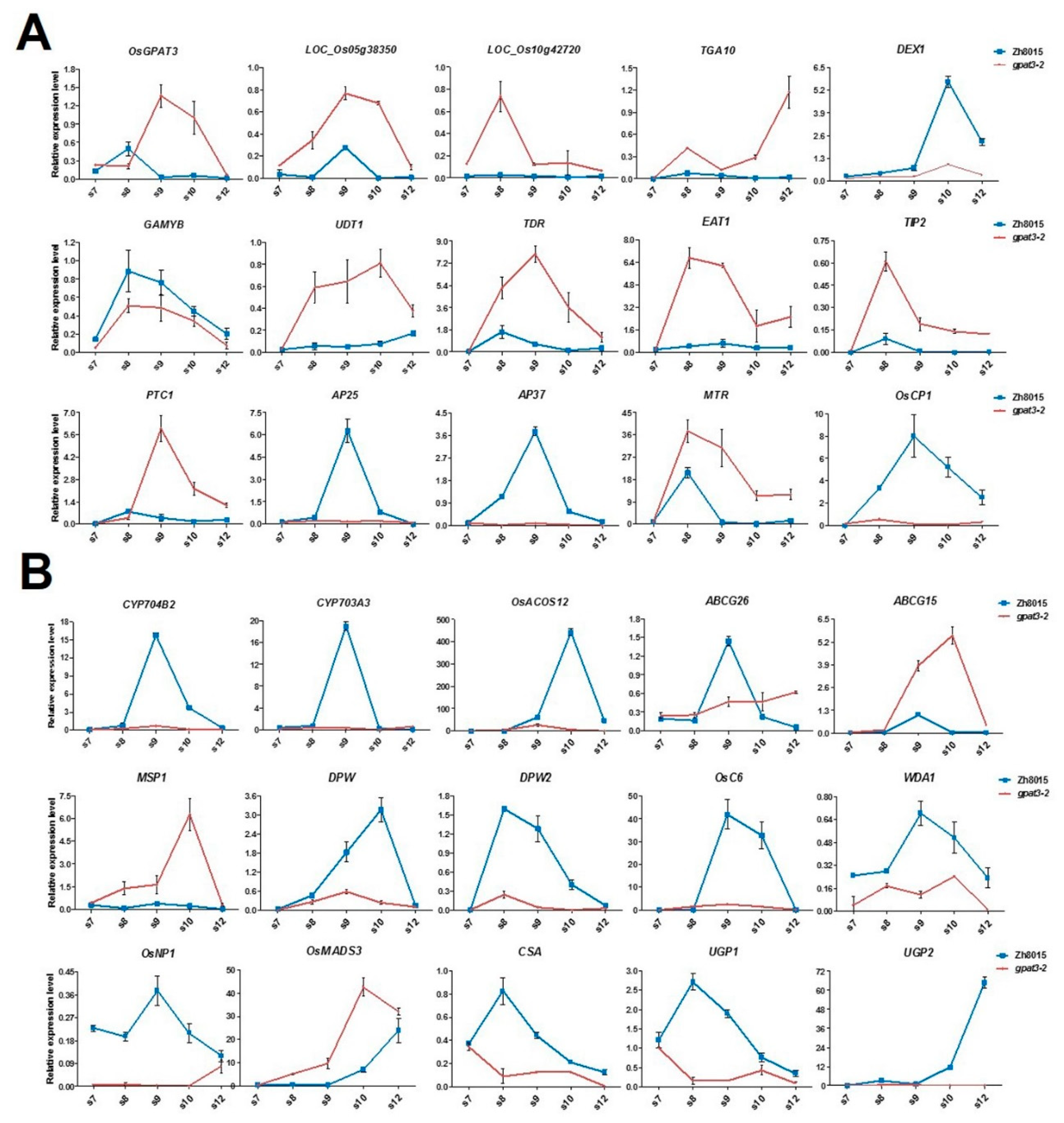
2.7. Mutation in OsGPAT3 Affects the Expression of Genes Involved in Both Tapetum PCD and Nutrient Metabolism
3. Discussion
3.1. OsGPAT3 Controls Ubisch Body Formation and Anther Development in Rice
3.2. Loss of Function of OsGPAT3 Causes Abnormal PCD of Both Anther Wall and Pollen Grains
3.3. Proposed Functions of OsGPAT3 in Rice Male Reproductive Development
4. Materials and Methods
4.1. Mutant Material and Plant Growth Conditions
4.2. Identification of Mutant Anther and Pollen Development
4.3. TUNEL Assay
4.4. Molecular Cloning of Osgpat3-2
4.5. Complementation of the gpat3-2 Mutant
4.6. Vector Construction for CRISPR/Cas9-Mediated Mutation
4.7. RNA Extraction, First-Strand Synthesis, and qPCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Goldberg, R.B.; Beals, T.P.; Sanders, P.M. Anther development: Basic principles and practical applications. Plant Cell 1993, 5, 1217–1229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.B.; Liang, W.Q. In Pushing the boundaries of scientific research: 120 years of addressing global issue. Science 2016, 351, 1223. [Google Scholar] [CrossRef]
- Ma, H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 2005, 56, 393–434. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.Z. Control of anther cell differentiation: A teamwork of receptor-like kinases. Sex. Plant Reprod. 2009, 22, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Wilson, Z.A.; Song, J.; Taylor, B.; Yang, C. The final split: The regulation of anther dehiscence. J. Exp. Bot. 2011, 62, 1633–1649. [Google Scholar] [CrossRef] [PubMed]
- Walbot, V.; Egger, R.L. Pre-meiotic anther development: Cell fate specification and differentiation. Annu. Rev. Plant Biol. 2016, 67, 365–395. [Google Scholar] [CrossRef]
- Scott, R.J.; Spielman, M.; Dickinson, H.G. Stamen structure and function. Plant Cell 2004, 16, S46–S60. [Google Scholar] [CrossRef]
- Matsui, T.; Omasa, K.; Horie, T. Mechanism of anther dehiscence in rice (Oryza sativa L.). Ann. Bot. 1999, 84, 501–506. [Google Scholar] [CrossRef]
- Wu, H.M.; Cheun, A.Y. Programmed cell death in plant reproduction. Plant Mol. Biol. 2000, 44, 267–281. [Google Scholar] [CrossRef]
- Parish, R.W.; Li, S.F. Death of a tapetum: A programme of developmental altruism. Plant Sci. 2010, 178, 73–89. [Google Scholar] [CrossRef]
- Jung, K.H.; Han, M.J.; Lee, Y.S.; Kim, Y.W.; Hwang, I.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; An, G. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 2015, 17, 2705–2722. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, D.S.; Liu, H.S.; Yin, C.S.; Li, X.X.; Liang, W.Q.; Yuan, Z.; Xu, B.; Chu, H.W.; Wang, J.; et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 2006, 18, 2999–3014. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Qu, L.J. Meiotic and mitotic cell cycle mutants involved in gametophyte development in Arabidopsis. Mol. Plant. 2008, 1, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Wilson, Z.A.; Zhang, D.B. From Arabidopsis to rice: Pathways in pollen development. J. Exp. Bot. 2009, 60, 1479–1492. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.X.; Cui, M.H.; Yang, L.; Kim, Y.J.; Zhang, D.B. Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci. 2015, 20, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Gavrieli, Y.; Sherma, Y.; Ben-Sasson, S.A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 1992, 119, 493–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varnier, A.L.; Mazeyrat-Gourbeyre, F.; Sangwan, R.S.; Clément, C. Programmed cell death progressively models the development of anther sporophytic tissues from the tapetum and is triggered in pollen grains during maturation. J. Struct Biol. 2005, 152, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.S.; Liang, W.Q.; Yuan, Z.; Li, N.; Shi, J.; Wang, J.; Liu, Y.M.; Yu, W.J.; Zhang, D.B. Tapetum Degeneration Retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol. Plant 2008, 1, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.N.; Li, H.Y.; Chen, L.B.; Xie, M.; Wang, F.P.; Chen, Y.L.; Liu, Y.G. A novel rice bHLH transcription factor, DTD, acts coordinately with tdr in controlling tapetum function and pollen development. Mol. Plant 2013, 6, 1715–1718. [Google Scholar] [CrossRef] [PubMed]
- Niu, N.N.; Liang, W.Q.; Yang, X.J.; Jin, W.L.; Wilson, Z.A.; Hu, J.P.; Zhang, D.B. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 2013, 4, 1445. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.Z.; Yu, J.; Cheng, X.W.; Zong, X.; Xu, J.; Chen, M.J.; Li, Z.Y.; Zhang, D.B.; Liang, W.Q. The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell 2014, 26, 1512–1524. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.S.; Li, M.J.; Sun-Ben, K.M.; Ho, Y.C.; Lin, Y.J.; Chuang, M.H.; Hsing, H.X.; Lien, Y.C.; Yang, H.T.; Chang, H.C.; et al. The bHLH142 transcription factor coordinates with tdr1 to modulate the expression of eat1 and regulate pollen development in rice. Plant Cell 2014, 26, 2486–2504. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.S.; Li, M.J.; Lin, Y.J.; Hsing, H.X.; Yang, T.T.; Chen, T.K.; Jhong, C.M.; Ku, M.S. Tightly controlled expression of bHLH142 is essential for timely tapetal programmed cell death and pollen development in rice. Front. Plant Sci. 2017, 8, 1258. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, R.; Khurana, R.; Malik, N.; Badoni, S.; Parida, S.K.; Kapoor, S.; Tyagi, A.K. bHLH142 regulates various metabolic pathway-related genes to affect pollen development and anther dehiscence in rice. Sci. Rep. 2017, 7, 43397. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, M.; Inukai, Y.; Ueguchi-Tanaka, M.; Itoh, H.; Izawa, T.; Kobayashi, Y.; Hattori, T.; Miyao, A.; Hirochika, H.; Ashikari, M.; et al. Loss-of-function mutations of the rice GAMYB gene impair α-amylase expression in aleurone and flower development. Plant Cell 2004, 16, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Aya, K.; Ueguchi-Tanaka, M.; Kondo, M.; Hamada, K.; Yano, K.; Nishimura, M.; Matsuoka, M. Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 2009, 21, 1453–1472. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.H.; Bao, W.J.; Liang, W.Q.; Yin, J.Y.; Zhang, D.B. Identification of gamyb-4 and analysis of the regulatory role of GAMYB in rice anther development. J. Integr. Plant Biol. 2010, 52, 670–678. [Google Scholar] [CrossRef]
- Li, H.; Yuan, Z.; Vizcay-Barrena, G.; Yang, C.Y.; Liang, W.Q.; Zong, J.; Wilson, Z.A.; Zhang, D.B. PERSISTENT TAPETAL CELL1 encodes a phd-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 2011, 156, 615–630. [Google Scholar] [CrossRef]
- Chen, Z.S.; Liu, X.F.; Wang, D.H.; Chen, R.; Zhang, X.L.; Xu, Z.H.; Bai, S.N. Transcription factor OsTGA10 is a target of the MADS protein OsMADS8 and is required for tapetum development. Plant Physiol. 2018, 176, 819–835. [Google Scholar] [CrossRef]
- Yi, J.; Moon, S.; Lee, Y.S.; Zhu, L.; Liang, W.Q.; Zhang, D.B.; Jung, K.H.; An, G. Defective Tapetum Cell Death 1 (DTC1) regulates ROS levels by binding to metallothionein during tapetum degeneration. Plant Physiol. 2016, 170, 1611–1623. [Google Scholar] [CrossRef]
- Yu, J.; Meng, Z.; Liang, W.Q.; Behera, S.; Kudla, J.; Tucker, M.R.; Luo, Z.J.; Chen, M.J.; Xu, D.W.; Zhao, G.C.; et al. A rice Ca2+ binding protein is required for tapetum function and pollen formation. Plant Physiol. 2016, 172, 1772–1786. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.X.; Liang, W.Q.; Hu, J.P.; Zhang, D.B. MTR1 encodes a secretory fasciclin glycoprotein required for male reproductive development in rice. Dev. Cell 2012, 22, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, Y.X.; Song, S.F.; Deng, H.F.; Li, N.; Fu, X.Q.; Chen, G.H.; Yuan, L.P. An anther development F-box (ADF) protein regulated by tapetum degeneration retardation (TDR) controls rice anther development. Planta 2015, 241, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Li, X.W.; Gao, X.Q.; Wei, Y.; Deng, L.; Ouyang, Y.D.; Chen, G.X.; Li, X.H.; Zhang, Q.F.; Wu, C.Y. Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-Box adenosine 5′-triphosphate-dependent rna helicases regulates tapetum degeneration. Plant Cell 2011, 23, 1416–1434. [Google Scholar] [CrossRef] [PubMed]
- Blackmore, S.; Wortley, A.H.; Skvarla, J.J.; Rowley, J.R. Pollen wall development in flowering plants. New Phytol. 2007, 174, 483–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.B.; Shi, J.X.; Yang, X. Role of lipid metabolism in plant pollen Exine development. Subcell Biochem. 2016, 86, 315–337. [Google Scholar] [CrossRef]
- Li, H.; Pinot, F.; Sauveplane, V.; Werckreichhart, D.; Diehl, P.; Schreiber, L.; Franke, R.; Zhang, P.; Chen, L.; Gao, Y.W.; et al. Cytochrome P450 family member CYP704B2 catalyzes the v-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 2010, 22, 173–190. [Google Scholar] [CrossRef]
- Yang, X.J.; Wu, D.; Shi, J.X.; Yi, H.; Pinot, F.; Grausem, B.; Yin, C.S.; Zhu, L.; Chen, M.J.; Luo, Z.J.; et al. Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J. Integr. Plant Biol. 2014, 56, 979–994. [Google Scholar] [CrossRef]
- Yang, Z.F.; Zhang, Y.X.; Sun, L.P.; Zhang, P.P.; Liu, L.; Yu, P.; Xuan, D.D.; Xiang, X.J.; Wu, W.X.; Cao, L.Y.; et al. Identification of cyp703a3-3 and analysis of regulatory role of CYP703A3 in rice anther cuticle and pollen exine development. Gene 2018, 649, 63–73. [Google Scholar] [CrossRef]
- Shi, J.; Tan, H.X.; Yu, X.H.; Liu, Y.Y.; Liang, W.Q.; Ranathunge, K.; Franke, R.B.; Schreiber, L.; Wang, Y.J.; Kai, G.Y.; et al. Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 2011, 23, 2225–2246. [Google Scholar] [CrossRef]
- Xu, D.W.; Shi, J.X.; Rautengarten, C.; Yang, L.; Qian, X.L.; Uzair, M.L.; Zhu, L.L.; Luo, Q.L.; An, G.L.; Waßmann, F.L.; et al. Defective Pollen Wall 2 (DPW2) encodes an acyl transferase required for rice pollen development. Plant Physiol. 2017, 173, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Qin, P.; Tu, B.; Wang, Y.P.; Deng, L.C.; Quilichini, T.D.; Li, T.; Wang, H.; Ma, B.T.; Li, S.G. ABCG15 encodes an ABC transporter protein, and is essential for post-meiotic anther and pollen exine development in rice. Plant Cell Physiol. 2013, 54, 138–154. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Shi, J.X.; Zhao, G.C.; Zhang, D.B.; Liang, W.Q. Post-meiotic deficient anther1 (PDA1) encodes an ABC transporter required for the development of anther cuticle and pollen exine in rice. J. Plant Biol. 2013, 56, 59–68. [Google Scholar] [CrossRef]
- Zhao, G.C.; Shi, J.X.; Liang, W.Q.; Xue, F.Y.; Luo, Q.; Zhu, L.; Qu, G.R.; Chen, M.J.; Schreiber, L.; Zhang, D.B. Two ATP Binding cassette G transporters, rice ATP binding cassette G26 and ATP binding cassette G15, collaboratively regulate rice male reproduction. Plant Physiol. 2015, 169, 2064–2079. [Google Scholar] [CrossRef] [PubMed]
- Chang, Z.Y.; Jin, M.N.; Yan, W.; Chen, H.; Qiu, S.J.; Fu, S.; Xia, J.X.; Liu, Y.C.; Chen, Z.F.; Wu, J.X.; et al. The ATP-binding cassette (ABC) transporter OsABCG3 is essential for pollen development in rice. Rice 2018, 11, 58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.S.; Liang, W.Q.; Yin, C.S.; Zong, J.; Gu, F.W.; Zhang, D.B. OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 2010, 154, 149–162. [Google Scholar] [CrossRef]
- Jung, K.H.; Han, M.J.; Lee, D.Y.; Lee, Y.S.; Schreiber, L.; Franke, R.; Faust, A.; Yephremov, A.; Saedler, H.; Kim, Y.W.; et al. Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 2006, 18, 3015–3032. [Google Scholar] [CrossRef]
- Li, Y.L.; Li, D.D.; Guo, Z.L.; Shi, Q.S.; Xiong, S.X.; Zhang, C.; Zhu, J.; Yang, Z.N. OsACOS12, an orthologue of Arabidopsis acyl-CoA synthetase5, plays an important role in pollen exine formation and anther development in rice. BMC Plant Biol. 2016, 16, 256. [Google Scholar] [CrossRef]
- Yang, X.J.; Liang, W.Q.; Chen, M.J.; Zhang, D.B.; Zhao, X.X.; Shi, J.X. Rice fatty acyl-CoA synthetase OsACOS12 is required for tapetum programmed cell death and male fertility. Planta 2017, 246, 105–122. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, S.; Shi, J.X.; Yu, J.; Zhu, L.; Yang, X.J.; Zhang, D.B.; Liang, W.Q. Rice No Pollen 1 (NP1) is required for anther cuticle formation and pollen exine patterning. Plant J. 2017, 91, 263–277. [Google Scholar] [CrossRef]
- Zhu, X.L.; Yu, J.; Shi, J.X.; Tohge, T.; Fernie, A.R.; Meir, S.; Aharoni, A.R.; Xu, D.W.; Zhang, D.B.; Liang, W.Q. The polyketide synthase OsPKS2 is essential for pollen exine and Ubisch body patterning in rice. J. Integr. Plant Biol. 2017, 59, 612–628. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.; Liu, M.X.; Xiao, Q.; Wang, T.; Chen, D.; Luo, T.; Yuan, G.Q.; Li, Q.; Zhu, J.; Liang, Y.Y.; et al. OsPKS2 is required for rice male fertility by participating in pollen wall formation. Plant Cell Rep. 2018, 37, 759–773. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.A.; Lee, D.P. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 2004, 43, 134–176. [Google Scholar] [CrossRef]
- Cao, J.; Li, J.L.; Li, D.; Tobin, J.F.; Gimeno, R.E. Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 19695–19700. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Reue, K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E1195–E1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lung, S.C.; Weselake, R.J. Diacylglycerol acyltransferase: A key mediator of splant triacylglycerol synthesis. Lipids 2006, 41, 1073–1088. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Beisson, F.; Koo, A.J.; Molina, I.; Pollard, M.; Ohlrogge, J. Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc. Natl. Acad. Sci. USA 2007, 104, 18339–18344. [Google Scholar] [CrossRef] [Green Version]
- Pollard, M.; Beisson, F.; Li, Y.; Ohlrogge, J.B. Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plant Sci. 2008, 13, 236–246. [Google Scholar] [CrossRef]
- Beisson, F.; Li-Beisson, Y.; Pollard, M. Solving the puzzles of cutin and suberin polymer biosynthesis. Curr. Opin. Plant Biol. 2012, 15, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Simpson, J.P.; Li-Beisson, Y.; Beisson, F.; Pollard, M.; Ohlrogge, J.B. A land-plant-specific glycerol-3-phosphate acyltransferase family in Arabidopsis: Substrate specificity, sn-2 preference, and evolution. Plant Physiol. 2012, 160, 638–652. [Google Scholar] [CrossRef]
- Zheng, Z.; Xia, Q.; Dauk, M.; Shen, W.; Selvaraj, G.; Zou, J. Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell 2003, 15, 1872–1887. [Google Scholar] [CrossRef] [PubMed]
- Petit, J.; Bres, C.; Mauxion, J.P.; Tai, F.W.; Martin, L.B.; Fich, E.A.; Joubes, J.; Rose, J.K.; Domergue, F.; Rothan, C. The glycerol-3-phosphate acyltransferase GPAT6 from tomato plays a central role in fruit cutin biosynthesis. Plant Physiol. 2016, 171, 894–913. [Google Scholar] [CrossRef] [PubMed]
- Men, X.; Shi, J.X.; Liang, W.Q.; Zhang, Q.F.; Lian, G.B.; Quan, S.; Zhu, L.; Luo, Z.J.; Chen, M.J.; Zhang, D.B. Glycerol-3-Phosphate Acyltransferase 3 (OsGPAT3) is required for anther development and male fertility in rice. J. Exp. Bot. 2017, 68, 513–526. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Wu, S.W.; Li, Z.W.; Zhou, Y.; Zhang, D.F.; Dong, Z.Y.; An, X.L.; Zhu, T.T.; Zhang, S.M.; Liu, S.S.; et al. Map-based cloning and characterization of Zea mays male sterility33 (ZmMs33) gene, encoding a glycerol-3-phosphate acyltransferase. Theor. Appl. Genet. 2018, 131, 1363–1378. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.B.; Wilson, Z.A. Stamen specification and anther development in rice. Chin. Sci. Bull. 2009, 54, 2342–2353. [Google Scholar] [CrossRef]
- Zhang, D.B.; Luo, X.; Zhu, L. Cytological analysis and genetic control of rice anther development. J. Genet. Genom. 2011, 38, 379–390. [Google Scholar] [CrossRef]
- Sun, L.P.; Zhang, Y.X.; Zhang, P.P.; Yang, Z.F.; Zhou, X.X.; Xuan, D.D.; Li, Z.H.; Wu, W.X.; Zhan, X.D.; Shen, X.H.; et al. Morphogenesis and Gene Mapping of deformed interior floral organ 1 (difo1), a novel mutant associated with floral organ development in rice. Plant Mol. Biol. Rep. 2017, 35, 330–344. [Google Scholar] [CrossRef]
- Shen, Y.J.; Jiang, H.; Jin, J.P.; Zhang, Z.B.; Xi, B.; He, Y.Y.; Wang, G.; Wang, C.; Qian, L.; Li, X.; et al. Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol. 2004, 135, 1198–1205. [Google Scholar] [CrossRef]
- Nonomura, K.I.; Miyoshi, K.; Eiguchi, M.; Suzuki, T.; Miyao, A.; Hirochika, H.; Kurata, N. The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 2003, 15, 1728–1739. [Google Scholar] [CrossRef]
- Hu, L.F.; Liang, W.Q.; Yin, C.S.; Cui, X.; Zong, J.; Wang, X.; Hu, J.P.; Zhang, D.B. Rice MADS3 regulates ros homeostasis during late anther development. Plant Cell 2011, 23, 515–533. [Google Scholar] [CrossRef]
- Ariizumi, T.; Toriyama, K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 2011, 62, 437–460. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.F.; Ho, T.H.; Wu, C.F.; Ho, S.L.; Yeh, R.H.; Lu, C.A.; Chen, P.W.; Yu, L.C.; Chao, A.; Yu, S.M. Convergent starvation signals and hormone crosstalk in regulating nutrient mobilization upon germination in cereals. Plant Cell 2012, 24, 2857–2873. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Y.X.; Liu, L.; Liu, Q.E.; Bi, Z.Z.; Yu, N.; Cheng, S.H.; Cao, L.Y. Fine mapping of the lesion mimic and early senescence 1 (lmes1) in rice (Oryza sativa. L). Plant Physiol. Biochem. 2014, 80, 300–307. [Google Scholar] [CrossRef]
- Chen, D.H.; Ronald, P.C. A rapid DNA minipreparation method suitable for AFLP and other PCR applications. Plant Mol. Biol. Rep. 1999, 17, 53–57. [Google Scholar] [CrossRef]
- Wu, W.X.; Zheng, X.M.; Chen, D.; Zhang, Y.X.; Ma, W.W.; Zhang, H.; Sun, L.P.; Yang, Z.F.; Zhao, C.D.; Zhan, X.D.; et al. OsCOL16, encoding a CONSTANS-like protein, represses flowering by up-regulating Ghd7 expression in rice. Plant Sci. 2017, 260, 60–69. [Google Scholar] [CrossRef]
- Miao, J.; Guo, D.; Zhang, J.; Huang, Q.; Qin, G.; Zhang, X.; Wan, J.; Gu, H.; Qu, L.J. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013, 23, 1233–1236. [Google Scholar] [CrossRef]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef]
- Zhang, H.; Liang, W.Q.; Yang, X.J.; Luo, X.; Jiang, N.; Ma, H.; Zhang, D.B. Carbon Starved Anther encodes a myb domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 2010, 22, 672–689. [Google Scholar] [CrossRef]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, L.; Xiang, X.; Yang, Z.; Yu, P.; Wen, X.; Wang, H.; Abbas, A.; Muhammad Khan, R.; Zhang, Y.; Cheng, S.; et al. OsGPAT3 Plays a Critical Role in Anther Wall Programmed Cell Death and Pollen Development in Rice. Int. J. Mol. Sci. 2018, 19, 4017. https://doi.org/10.3390/ijms19124017
Sun L, Xiang X, Yang Z, Yu P, Wen X, Wang H, Abbas A, Muhammad Khan R, Zhang Y, Cheng S, et al. OsGPAT3 Plays a Critical Role in Anther Wall Programmed Cell Death and Pollen Development in Rice. International Journal of Molecular Sciences. 2018; 19(12):4017. https://doi.org/10.3390/ijms19124017
Chicago/Turabian StyleSun, Lianping, Xiaojiao Xiang, Zhengfu Yang, Ping Yu, Xiaoxia Wen, Hong Wang, Adil Abbas, Riaz Muhammad Khan, Yingxin Zhang, Shihua Cheng, and et al. 2018. "OsGPAT3 Plays a Critical Role in Anther Wall Programmed Cell Death and Pollen Development in Rice" International Journal of Molecular Sciences 19, no. 12: 4017. https://doi.org/10.3390/ijms19124017



