Hypothesis: Spontaneous Advent of the Prebiotic Translation System via the Accumulation of L-Shaped RNA Elements
Abstract
:1. Introduction
2. Results and Discussion
2.1. Shape of the Ancestral tRNA
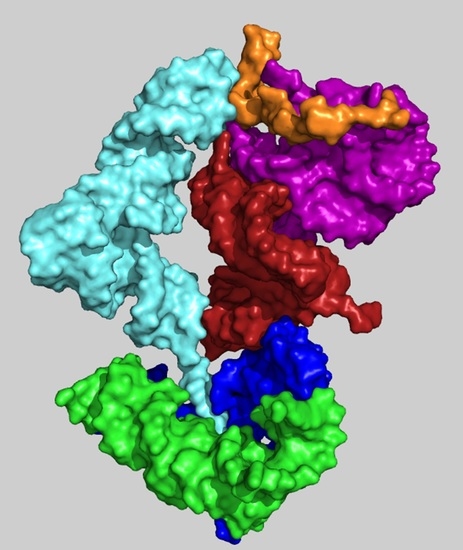
2.2. A Model of a Coded Proto-Ribosome
2.3. Spontaneous Formation of the Coded Proto-Ribosome Components
2.4. Evolutionary Path of the Translation System
3. Materials and Methods
4. Conclusion
Supplementary Materials
Acknowledgments
Conflicts of Interest
Abbreviations
| DPR | Dimeric Proto Ribosome |
| aa | Amino Acid |
| PTC | Peptidyl Transferase Center |
| LSU | Large Subunit of the Ribosome |
| SSU | Small Subunit of the Ribosome |
| AC | Anticodon |
| rRNA | Ribosomal RNA |
| A-tRNA | A-site tRNA |
| P-tRNA | P-site tRNA |
References
- Hopfield, J.J. Origin of the genetic code: A testable hypothesis based on tRNA structure, sequence and kinetic proofreading. Proc. Natl. Acad. Sci. USA 1978, 75, 4334–4338. [Google Scholar] [CrossRef] [PubMed]
- Möller, W.; Janssen, G.M. Statistical evidence for remnants of the primordial code in the acceptor stem of prokaryotic transfer RNA. J. Mol. Evol. 1992, 34, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Maizels, N.; Weiner, A.M. Phylogeny from function: Evidence from the molecular fossil record that tRNA originated in replication, not translation. Proc. Natl. Acad. Sci. USA 1994, 91, 6729–6734. [Google Scholar] [CrossRef] [PubMed]
- Schimmel, P.; Ribas de Pouplana, L. Transfer RNA: From minihelix to genetic code. Cell 1995, 81, 983–986. [Google Scholar] [CrossRef] [Green Version]
- Musier-Forsyth, K.; Schimmel, P. Atomic determinants for aminoacylation of RNA minihelices and relationship to genetic code. Acc. Chem. Res. 1999, 32, 368–375. [Google Scholar] [CrossRef]
- Di Giulio, M. The origin of the tRNA molecule: Implications for the origin of protein synthesis. J. Theor. Biol. 2004, 226, 89–93. [Google Scholar] [CrossRef]
- Kuhn, H.; Waser, J. Evolution of early mechanisms of translation of genetic information into polypeptides. Nature 1982, 298, 585–586. [Google Scholar] [CrossRef]
- Schimmel, P.; Henderson, B. Possible role of aminoacyl-RNA complexes in noncoded peptide synthesis and origin of coded synthesis. Proc. Natl. Acad. Sci. USA. 1994, 91, 11283–11286. [Google Scholar] [CrossRef]
- Lehmann, J. Physico-chemical constraints connected with the coding properties of the genetic system. J. Theor. Biol. 2000, 202, 129–144. [Google Scholar] [CrossRef]
- Purohit, P.; Stern, S. Interactions of a small RNA with antibiotic and RNA ligands of the 30S subunit. Nature 1994, 370, 659–662. [Google Scholar] [CrossRef]
- Agmon, I.; Bashan, A.; Yonath, A. On ribosome conservation and evolution. Isr. J. Ecol. Evol. 2006, 52, 359–374. [Google Scholar] [CrossRef]
- Anderson, R.M.; Kwon, M.; Strobel, S.A. Toward ribosomal RNA catalytic activity in the absence of protein. J. Mol. Evol. 2007, 64, 472–483. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.F.; Lee, J.C.; Gutell, R.R.; Hartman, H. The origin and evolution of the ribosome. Biol. Direct. 2008, 3, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokov, K.; Steinberg, S.V. A hierarchical model for evolution of 23S ribosomal RNA. Nature 2009, 457, 977–980. [Google Scholar] [CrossRef] [PubMed]
- Agmon, I. The dimeric proto-ribosome: Structural details and implications on the origin of life. Int. J. Mol. Sci. 2009, 10, 2921–2934. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, C.; Mohan, S.; Kalahar, B.K.; Williams, L.D. Peeling the onion: Ribosomes are ancient molecular fossils. Mol. Biol. Evol. 2009, 26, 2415–2425. [Google Scholar] [CrossRef] [PubMed]
- Petrov, A.S.; Gulen, B.; Norris, A.M.; Kovacs, N.A.; Bernier, C.R.; Lanier, K.A.; Fox, G.E.; Harvey, S.C.; Wartell, R.M.; Hud, N.V.; et al. History of the ribosome and the origin of translation. Proc. Natl. Acad. Sci. USA 2015, 112, 15396–15401. [Google Scholar] [CrossRef] [PubMed]
- Agmon, I.C. Could a proto-ribosome emerge spontaneously in the prebiotic world? Molecules 2016, 21, E1701. [Google Scholar] [CrossRef] [PubMed]
- Agmon, I.; Bashan, A.; Zarivach, R.; Yonath, A. Symmetry at the active site of the ribosome: Structural and functional implications. Biol. Chem. 2005, 386, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Agmon, I. Sequence complementarity at the ribosomal Peptidyl Transferase Centre implies self-replicating origin. FEBS Lett. 2017, 591, 3252–3258. [Google Scholar] [CrossRef] [PubMed]
- Yusupov, M.M.; Yusupova, G.Z.; Baucom, A.; Lieberman, K.; Earnest, T.N.; Cate, J.H.; Noller, H.F. Crystal structure of the ribosome at 5.5 A resolution. Science 2001, 292, 883–896. [Google Scholar] [CrossRef] [PubMed]
- Orgel, L.E. The origin of polynucleotide-directed protein synthesis. J. Mol. Evol. 1989, 29, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Nissen, P.; Ippolito, J.A.; Ban, N.; Moore, P.B.; Steitz, T.A. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. USA 2001, 98, 4899–4903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Fredrick, K. Intersubunit Bridges of the Bacterial Ribosome. J. Mol. Biol. 2016, 428, 2146–2164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunkle, J.A.; Cate, J.H. Ribosome structure and dynamics during translocation and termination. Annu. Rev. Biophys. 2010, 39, 227–244. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [Green Version]
- Mast, C.B.; Schink, S.; Gerland, U.; Braun, D. Escalation of polymerization in a thermal gradient. Proc. Natl. Acad. Sci. USA 2013, 110, 8030–8035. [Google Scholar] [CrossRef] [Green Version]
- Briones, C.; Stich, M.; Manrubia, S.C. The dawn of the RNA World: Toward functional complexity through ligation of random RNA oligomers. RNA 2009, 15, 743–749. [Google Scholar] [CrossRef] [Green Version]
- Di Giulio, M. On the origin of the transfer RNA molecule. J. Theor. Biol. 1992, 159, 199–214. [Google Scholar] [CrossRef]
- Dick, T.P.; Schamel, W.A. Molecular evolution of transfer RNA from two precursor hairpins: Implications for the origin of protein synthesis. J. Mol. Evol. 1995, 1, 1–9. [Google Scholar] [CrossRef]
- Nagaswamy, U.; Fox, G.E. RNA ligation and the origin of tRNA. Orig. Life Evol. Biosph. 2003, 33, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Gutell, R.R.; Uhlenbeck, O.C. Folding of circularly permuted transfer RNAs. Science 1991, 254, 1361–1364. [Google Scholar] [CrossRef] [PubMed]
- Root-Bernstein, R.; Kim, Y.; Sanjay, A.; Burton, Z.F. tRNA evolution from the proto-tRNA minihelix world. Transcription 2016, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kowiatek, B.; Opron, K.; Burton, Z.F. Type-II tRNAs and Evolution of Translation Systems and the Genetic Code. Int. J. Mol. Sci. 2018, 19, 3257. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, A.; Nakashima, T.; Taniguchi, M.; Hosaka, H.; Kimura, M.; Tanaka, I. The three-dimensional structure of the RNA-binding domain of ribosomal protein L2; a protein at the peptidyl transferase center of the ribosome. EMBO J. 1999, 18, 1459–1467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovacs, N.A.; Petrov, A.S.; Lanier, K.A.; Williams, L.D. Frozen in Time: The History of Proteins. Mol. Biol. Evol. 2017, 34, 1252–1260. [Google Scholar] [CrossRef]
- Fayerverker, I.; Mor, T. On Code-Prompting Auto-Catalytic Sets and the Origins of Coded Life. In Proceedings of the 3rd International Conference on Complexity, Future Information Systems and Risk, Madeira, Portugal, 20–21 March 2018; pp. 53–63. [Google Scholar]
- Gavrilova, L.P.; Koteliansky, V.E.; Spirin, A.S. Ribosomal protein s12 and ‘non enzymatic’ translocation. FEBS Lett. 1974, 45, 324–328. [Google Scholar] [CrossRef]
- Krzyzaniak, A.; Barciszewski, J.; Sałański, P.; Jurczak, J. The non-enzymatic specific amino-acylation of transfer RNA at high pressure. Int. J. Biol. Macromol. 1994, 16, 153–158. [Google Scholar] [CrossRef]
- Prywes, N.; Blain, J.C.; Del Frate, F.; Szostak, J.W. Nonenzymatic copying of RNA templates containing all four letters is catalyzed by activated oligonucleotides. Elife 2016, 5, e17756. [Google Scholar] [CrossRef]
- Agmon, I.; Mor, T. A Model for the Emergence of Coded Life. In Proceedings of the Theory and Practice of Natural Computing, Mieres, Spain, 15–16 December 2015; pp. 97–108. [Google Scholar]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and Development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Cannone, J.J.; Subramanian, S.; Schnare, M.N.; Collett, J.R.; D’Souza, L.M.; Du, Y.; Feng, B.; Lin, N.; Madabusi, L.V.; Müller, K.M.; et al. The comparative RNAWeb CRWSite: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinform. 2002, 3, 1–31. [Google Scholar]






| A-Minor Interaction | Nucleotides Involved (Location) | Contemporary Role |
|---|---|---|
| A-DPR → P-DPR | A2598 → C2073:G2436 (H93 stem loop) (H74 stem) | Part of the GNRA interaction between A-, P-DPR [15] (Figure S1) |
| H69–71 bridging element → A-DPR [14] | A1966 → G2592:C2601 (H71–H67 loop) (H93 stem) | |
| H69–71 bridging element → h44–45 proto-SSU [24] | A1912 → C1407:G1494 (H69 stem loop) (h44 stem) | Part of the B2a intersubunit bridge [24] |
| h44–45 proto-SSU → H69–71 bridging element [24] | a. A1418 → G1948:C1958 (h44 stem) (H71 stem) b. A1483 → C1947:G1959 (h44 stem) (H71 stem) | Part of the B3 intersubunit bridge [24] and center of the intersubunit ratchet [25] |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agmon, I. Hypothesis: Spontaneous Advent of the Prebiotic Translation System via the Accumulation of L-Shaped RNA Elements. Int. J. Mol. Sci. 2018, 19, 4021. https://doi.org/10.3390/ijms19124021
Agmon I. Hypothesis: Spontaneous Advent of the Prebiotic Translation System via the Accumulation of L-Shaped RNA Elements. International Journal of Molecular Sciences. 2018; 19(12):4021. https://doi.org/10.3390/ijms19124021
Chicago/Turabian StyleAgmon, Ilana. 2018. "Hypothesis: Spontaneous Advent of the Prebiotic Translation System via the Accumulation of L-Shaped RNA Elements" International Journal of Molecular Sciences 19, no. 12: 4021. https://doi.org/10.3390/ijms19124021