Knockout of SlSBPASE Suppresses Carbon Assimilation and Alters Nitrogen Metabolism in Tomato Plants
Abstract
:1. Introduction
2. Results
2.1. Generation of Loss-of-Function Mutant of SlSBPASE Using CRISPR/Cas9 Gene-Editing System
2.2. Characterization of slsbpase Mutant Plants
2.3. Mutation of SlSBPASE Causes Severe Reductions in SBPase Activity, RuBP Regeneration and CO2 Assimilation Rate
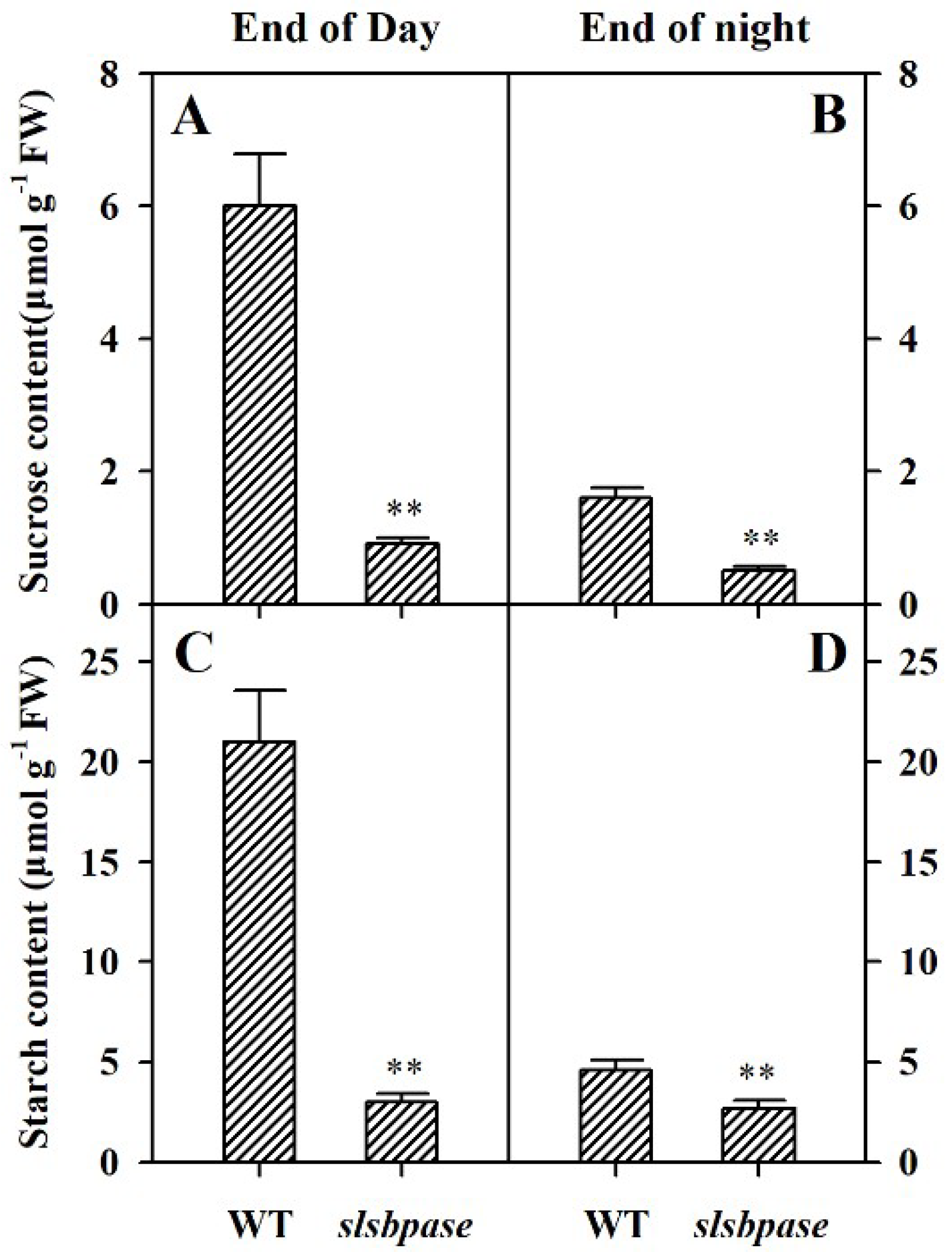
2.4. Mutation of SlSBPASE Dramatically Inhibits Biosynthesis of Sucrose and Starch
2.5. Mutation of SlSBPASE Reduces Night Respiration Rate
2.6. Mutation of SlSBPASE Decreases Levels of Protein and Amino Acids in Tomato Leaves
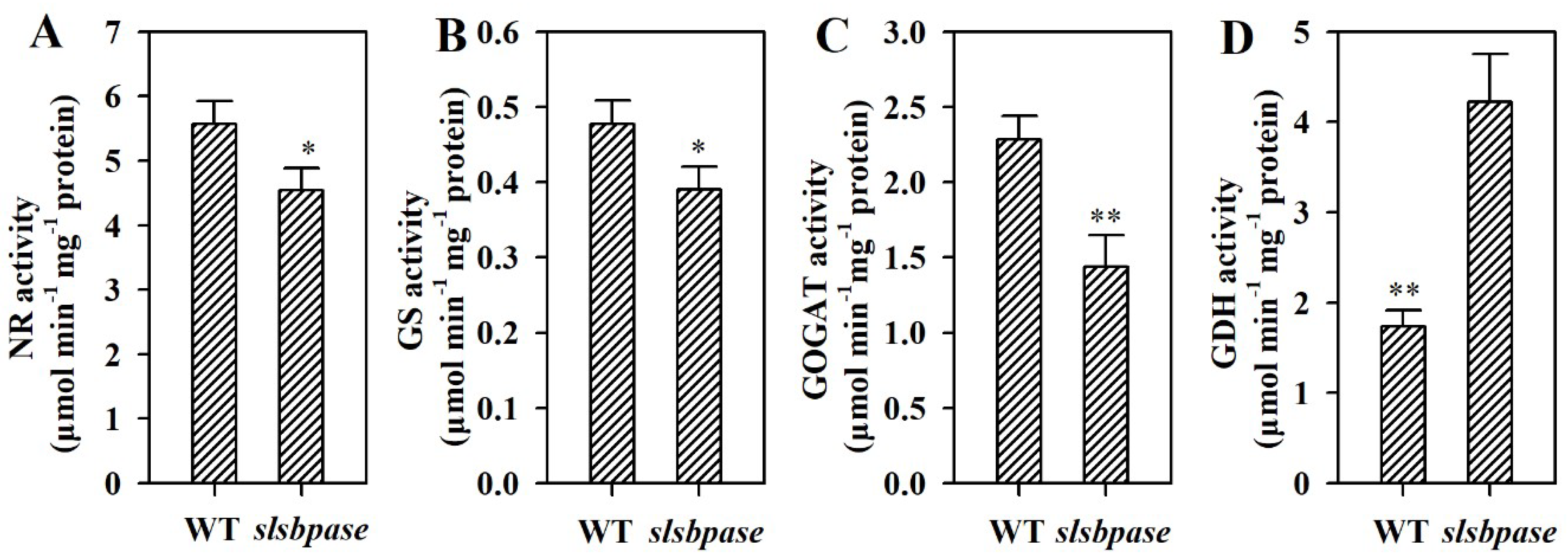
2.7. Mutation of SlSBPASE Alters Activities of Enzymes Involved in Nitrogen Metabolism
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Selection of Target Sequence and Plasmid Construction
4.3. Agrobacterium Tumefaciens-Mediated Transformation
4.4. Mutation Analysis of Transgenic Lines
4.5. Off-Target Analysis
4.6. Activities of SBPase, NR, GS, GOGAT and GDH
4.7. Measurement of Carbon Assimilation Rate and Night Respiration Rate
4.8. Determination of RuBP Level
4.9. Determination of Carbohydrate Level
4.10. Measurements of Protein and Amino Acids
4.11. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Geiger, D.R.; Servaites, J.C. Diurnal regulation of photosynthetic carbon metabolism in C3 plants. Annu. Rev. Plant Biol. 1994, 45, 235–256. [Google Scholar] [CrossRef]
- Raines, C.A.; Lloyd, J.C.; Dyer, T.A. New insights into the structure and function of sedoheptulose-1,7-bisphosphatase; an important but neglected Calvin cycle enzyme. J. Exp. Bot. 1999, 50, 1–8. [Google Scholar] [Green Version]
- Laing, W.A.; Stitt, M.; Heldt, H.W. Control of CO2 fixation. Changes in the activity of ribulosephosphate kinase and fructose- and sedoheptulose-bisphosphatase in chloroplasts. BBA Bioenerg. 1981, 637, 348–359. [Google Scholar] [CrossRef]
- Woodrow, I.E.; Murphy, D.J.; Latzko, E. Regulation of stromal sedoheptulose 1,7-bisphosphatase activity by pH and Mg2+ concentration. J. Biol. Chem. 1984, 259, 3791–3795. [Google Scholar]
- Breazeale, V.D.; Buchanan, B.B.; Wolosiuk, R.A. Chloroplast sedoheptulose-1,7-bisphosphatase: Evidence for regulation by the ferredoxin/thioredoxin system. Z. Naturforsch. C 1978, 33, 521–528. [Google Scholar] [CrossRef]
- Wirtz, W.; Stitt, M.; Heldt, H.W. Light activation of calvin cycle enzymes as measured in pea leaves. FEBS Lett. 1982, 142, 223–226. [Google Scholar] [CrossRef] [Green Version]
- Miyagawa, Y.; Tamoi, M.; Shigeoka, S. Overexpression of a cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase in tobacco enhances photosynthesis and growth. Nat. Biotechnol. 2001, 19, 965–969. [Google Scholar] [CrossRef]
- Lefebvre, S.; Lawson, T.; Fryer, M.; Zakhleniuk, O.V.; Lloyd, J.C.; Raines, C. Increased sedoheptulose-1, 7-bisphosphatase activity in transgenic tobacco plants stimulates photosynthesis and growth from an early stage in development. Plant Physiol. 2005, 138, 451–460. [Google Scholar] [CrossRef]
- Rosenthal, D.M.; Locke, A.M.; Khozaei, M.; Raines, C.A.; Long, S.P.; Ort, D.R. Over-expressing the C3 photosynthesis cycle enzyme sedoheptulose-1-7 bisphosphatase improves photosynthetic carbon gain and yield under fully open air CO2 fumigation (FACE). BMC Plant Biol. 2011, 11, 123. [Google Scholar] [CrossRef]
- Liu, X.L.; Yu, H.D.; Guan, Y.; Li, J.K.; Guo, F.Q. Carbonylation and loss-of-function analyses of SBPase reveal its metabolic interface role in oxidative stress, carbon assimilation and multiple aspects of growth and development in Arabidopsis. Mol. Plant 2012, 5, 1082–1099. [Google Scholar] [CrossRef]
- Driever, S.M.; Simkin, A.J.; Alotaibi, S.; Fisk, S.J.; Madgwick, P.J.; Sparks, C.A.; Jones, H.D.; Lawson, T.; Parry, M.A.J.; Raines, C.A. Increased SBPase activity improves photosynthesis and grain yield in wheat grown in greenhouse conditions. Philos. Trans. R. Soc. B 2017, 372, 20160384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stitt, M.; Matt, P.; Gibon, Y. Steps towards an integrated view of nitrogen metabolism. J. Exp. Bot. 2002, 53, 959–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabuchi, M.; Abiko, T.; Yamaya, T. Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.). J. Exp. Bot. 2007, 58, 2319–2327. [Google Scholar] [CrossRef] [PubMed]
- Stitt, M. Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol. 1999, 2, 178–186. [Google Scholar] [CrossRef]
- Gao, L.; Lu, Z.; Ding, L.; Guo, J.; Wang, M.; Ling, N.; Guo, S.; Shen, Q. Role of aquaporins in determining carbon and nitrogen status in higher plants. Int. J. Mol. Sci. 2018, 19, 35. [Google Scholar] [CrossRef] [PubMed]
- Gibon, Y.; Pyl, E.T.; Sulpice, R.; Lunn, J.E.; HÖhne, M.; GÜnther, M.; Stitt, M. Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ. 2009, 32, 859–874. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Li, H.; Jiao, J.; Li, D.; Zhou, L.; Wan, J.; Li, Y. Reduction in SBPase activity by antisense RNA in transgenic rice plants: Effect on photosynthesis, growth and biomass allocation at different nitrogen levels. J. Plant Biol. 2009, 52, 382–394. [Google Scholar] [CrossRef]
- Alagoz, Y.; Gurkok, T.; Zhang, B.; Unver, T. Manipulating the biosynthesis of bioactive compound alkaloids for next-generation metabolic engineering in opium poppy using CRISPR-Cas 9 genome editing technology. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef]
- Liu, X.; Wu, S.; Xu, J.; Sui, C.; Wei, J. Application of CRISPR/Cas9 in plant biology. Acta Pharm. Sin. B 2017, 7, 292–302. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Chen, L.; Li, R.; Zhao, R.; Yang, M.; Sheng, J.; Shen, L. Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 2017, 65, 8674–8682. [Google Scholar] [CrossRef]
- Li, T.; Yang, X.; Yu, Y.; Si, X.; Zhai, X.; Zhang, H.; Dong, W.; Gao, C.; Xu, C. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Patron, N.J.; Nekrasov, V. Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 2015, 32, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Wang, M.; Zhang, S.; Ai, X. Changes in SBPase activity influence photosynthetic capacity, growth and tolerance to chilling stress in transgenic tomato plants. Sci. Rep. 2016, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Wang, M.; Zhang, S. Overexpression of a Calvin cycle enzyme SBPase improves tolerance to chilling-induced oxidative stress in tomato plants. Sci. Hortic. 2017, 214, 27–33. [Google Scholar] [CrossRef]
- Harrison, E.P.; Olcer, H.; Lloyd, J.C.; Long, S.P.; Raines, C.A. Small decreases in SBPase cause a linear decline in the apparent RuBP regeneration rate but do not affect Rubisco carboxylation capacity. J. Exp. Bot. 2001, 52, 1779–1784. [Google Scholar] [CrossRef] [PubMed]
- Harrison, E.P.; Willingham, N.M.; Lloyd, J.C.; Raines, C.A. Reduced sedoheptulose-1,7-bisphosphatase levels in transgenic tobacco lead to decreased photosynthetic capacity and altered carbohydrate accumulation. Planta 1998, 204, 27–36. [Google Scholar] [CrossRef]
- Olçer, H.; Lloyd, J.C.; Raines, C.A. Photosynthetic capacity is differentially affected by reductions in sedoheptulose-1,7-bisphosphatase activity during leaf development in transgenic tobacco plants. Plant Physiol. 2001, 125, 982–989. [Google Scholar] [CrossRef]
- Lawson, T.; Bryant, B.; Lefebvre, S.; Lloyd, J.C.; Raines, C.A. Decreased SBPase activity alters growth and development in transgenic tobacco plants. Plant Cell Environ. 2006, 29, 48–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raines, C.A.; Harrison, E.P.; Lloyd, J.C. Investigating the role of the thiol-regulated enzyme sedoheptulose-1, 7-bisphosphatase in the control of photosynthesis. Physiol. Plant. 2000, 110, 303–308. [Google Scholar] [CrossRef]
- Simkin, A.J.; Lopez-Calcagno, P.E.; Davey, P.A.; Headland, L.R.; Lawson, T.; Timm, S.; Bauwe, H.; Raines, C.A. Simultaneous stimulation of sedoheptulose 1,7-bisphosphatase, fructose 1,6-bisphophate aldolase and the photorespiratory glycine decarboxylase-H protein increases CO2assimilation, vegetative biomass and seed yield in Arabidopsis. Plant Biotechnol. J. 2017, 15, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Han, Y.; Liu, G.; An, B.; Yang, J.; Yang, G.; Li, Y.; Zhu, Y. Overexpression of sedoheptulose-1, 7-bisphosphatase enhances photosynthesis and growth under salt stress in transgenic rice plants. Funct. Plant Biol. 2007, 34, 822–834. [Google Scholar] [CrossRef]
- Feng, L.; Wang, K.; Li, Y.; Tan, Y.; Kong, J.; Li, H.; Li, Y.; Zhu, Y. Overexpression of SBPase enhances photosynthesis against high temperature stress in transgenic rice plants. Plant Cell Rep. 2007, 26, 1635–1646. [Google Scholar] [CrossRef]
- Ding, F.; Wang, M.; Zhang, S. Sedoheptulose-1,7-bisphosphatase is involved in methyl jasmonate- and dark-induced leaf senescence in tomato plants. Int. J. Mol. Sci. 2018, 19, 3673. [Google Scholar] [CrossRef] [PubMed]
- Salvucci, M.E.; Ogren, W.L. The mechanism of Rubisco activase: Insights from studies of the properties and structure of the enzyme. Photosynth. Res. 1996, 47, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Raines, C.A.; Lloyd, J.C.; Willingham, N.M.; Potts, S.; Dyer, T.A. cDNA and gene sequences of wheat chloroplast sedoheptulose-1,7-bisphosphatase reveal homology with fructose-1,6-bisphosphatases. Eur. J. Biochem. 1992, 205, 1053–1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gütle, D.D.; Roret, T.; Müller, S.J.; Couturier, J.; Lemaire, S.D.; Hecker, A.; Dhalleine, T.; Buchanan, B.B.; Reski, R.; Einsle, O.; Jacquot, J.-P. Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories. Proc. Natl. Acad. Sci. USA 2016, 113, 6779–6784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bloom, A.J. Photorespiration and nitrate assimilation: A major intersection between plant carbon and nitrogen. Photosynth. Res. 2015, 123, 117–128. [Google Scholar] [CrossRef]
- Miflin, B.J.; Habash, D.Z. The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities. J. Exp. Bot. 2002, 53, 979–987. [Google Scholar] [CrossRef]
- Guo, Q.; Yoshida, Y.; Major, I.T.; Wang, K.; Sugimoto, K.; Kapali, G.; Havko, N.E.; Benning, C.; Howe, G.A. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E10768–E10777. [Google Scholar] [CrossRef]
- Xie, X.; Ma, X.; Zhu, Q.; Zeng, D.; Li, G.; Liu, Y.G. CRISPR-GE: A convenient software toolkit for CRISPR-based genome editing. Mol. Plant 2017, 10, 1246–1249. [Google Scholar] [CrossRef]
- Dan, Y.; Yan, H.; Munyikwa, T.; Dong, J.; Zhang, Y.; Armstrong, C.L. MicroTom—A high-throughput model transformation system for functional genomics. Plant Cell Rep. 2006, 25, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Sicher, R.C.; Bahr, J.T.; Jensen, R.G. Measurement of ribulose 1, 5-bisphosphate from spinach chloroplasts. Plant Physiol. 1979, 64, 876–879. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Song, W.; Sage, T.L.; DellaPenna, D. Tocopherols play a crucial role in low-temperature adaptation and phloem loading in Arabidopsis. Plant Cell 2006, 18, 2710–2732. [Google Scholar] [CrossRef] [PubMed]
- Stitt, M.; Lilley, R.; Gerhardt, R.; Heldt, H.W. Metabolite levels in specific cells and subcellular compartments of plant leaves. In Methods in Enzymology; Fleischer, S., Fleischer, B., Eds.; Academic Press: Cambridge, MA, USA, 1989; Volume 174, pp. 518–552. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
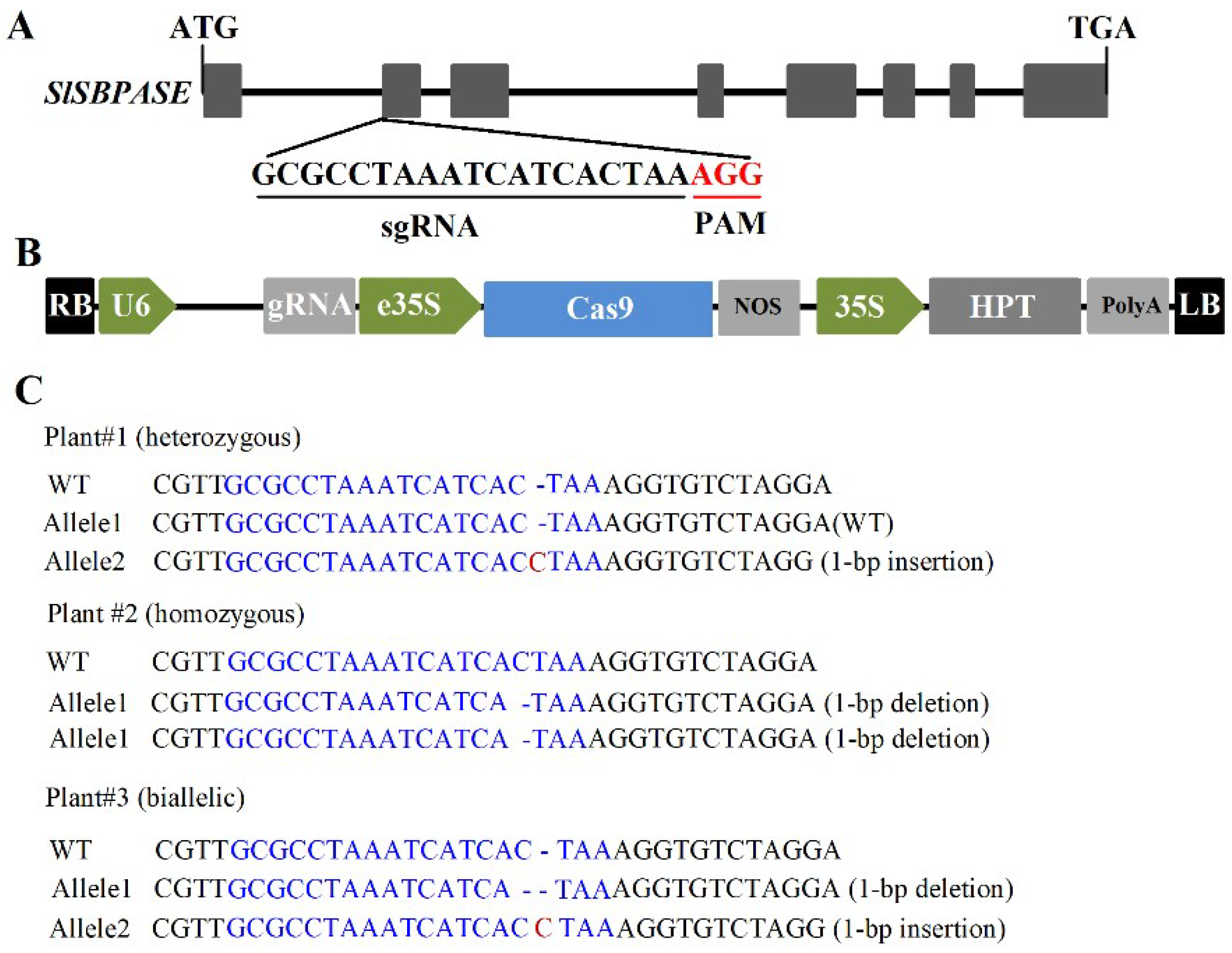




| Potential off-Target Sites | Primer Sequence (5′–3′) |
|---|---|
| Site 1 | F1: TGGGTCTGAATCATCACTAG |
| TGGGTCTGAATCATCACTAGAGG | R1: CTAGTGATGATTCAGACCCA |
| Site 2 | F2: AGTGCCTAAATCCTCAATAA |
| AGTGCCTAAATCCTCAATAAAGG | R2: TTATTGAGGATTTAGGCACT |
| Site 3 | F3: TGTGTCTAAATCATCATTCA |
| TGTGTCTAAATCATCATTCAAGG | R3: TGAATGATGATTTAGACACA |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, F.; Hu, Q.; Wang, M.; Zhang, S. Knockout of SlSBPASE Suppresses Carbon Assimilation and Alters Nitrogen Metabolism in Tomato Plants. Int. J. Mol. Sci. 2018, 19, 4046. https://doi.org/10.3390/ijms19124046
Ding F, Hu Q, Wang M, Zhang S. Knockout of SlSBPASE Suppresses Carbon Assimilation and Alters Nitrogen Metabolism in Tomato Plants. International Journal of Molecular Sciences. 2018; 19(12):4046. https://doi.org/10.3390/ijms19124046
Chicago/Turabian StyleDing, Fei, Qiannan Hu, Meiling Wang, and Shuoxin Zhang. 2018. "Knockout of SlSBPASE Suppresses Carbon Assimilation and Alters Nitrogen Metabolism in Tomato Plants" International Journal of Molecular Sciences 19, no. 12: 4046. https://doi.org/10.3390/ijms19124046




