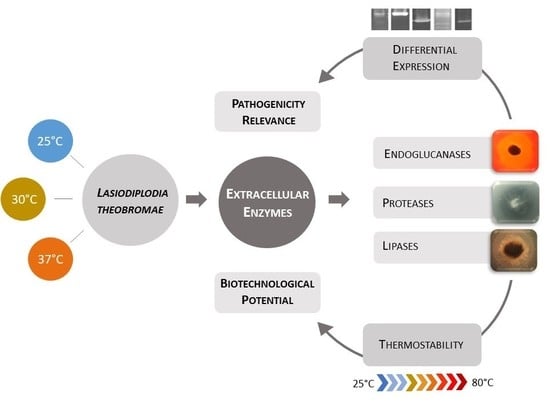
Lasiodiplodia theobromae as a Producer of Biotechnologically Relevant Enzymes
Abstract
:1. Introduction
2. Results
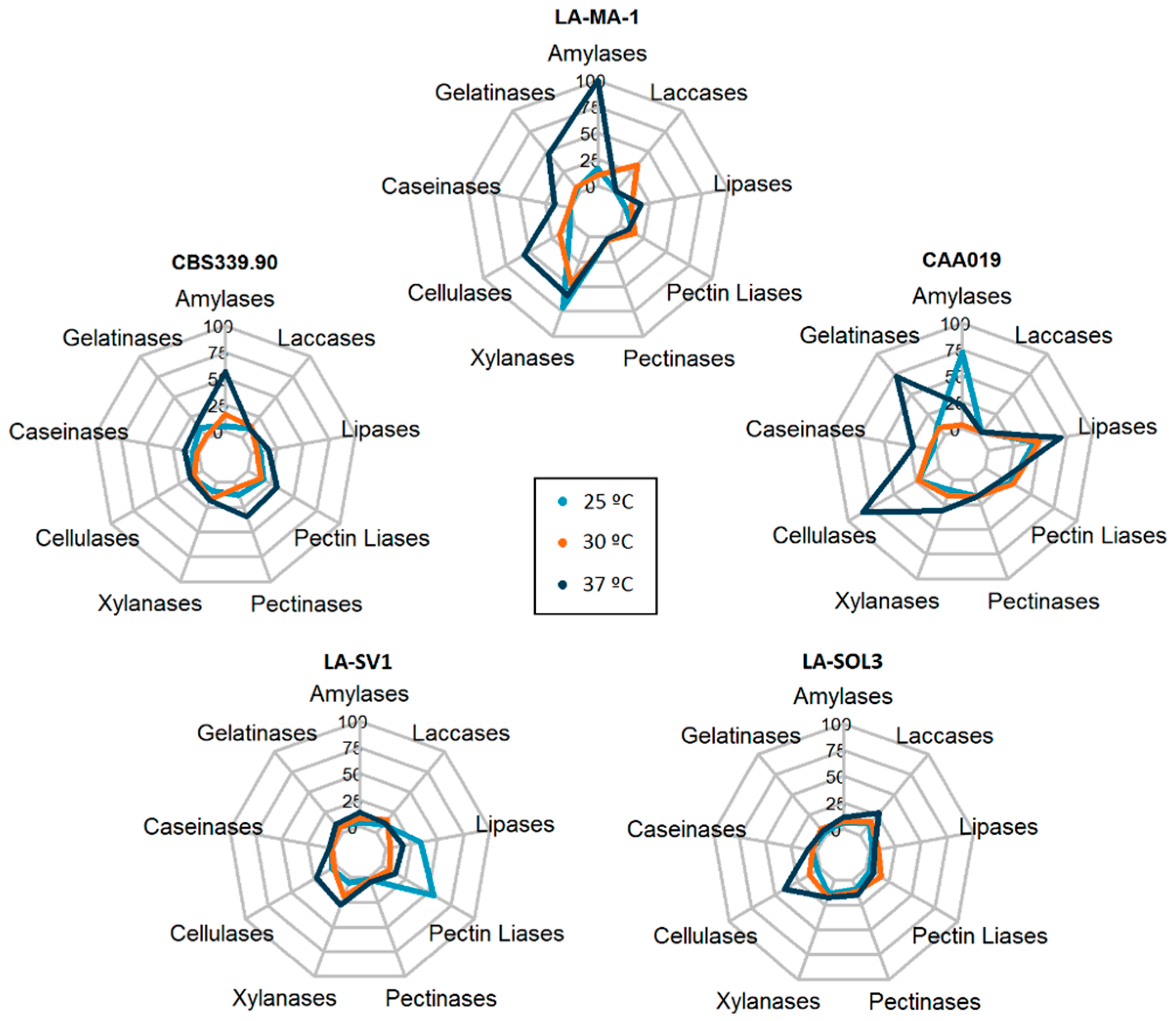
2.1. Extracellular Enzymatic Activity
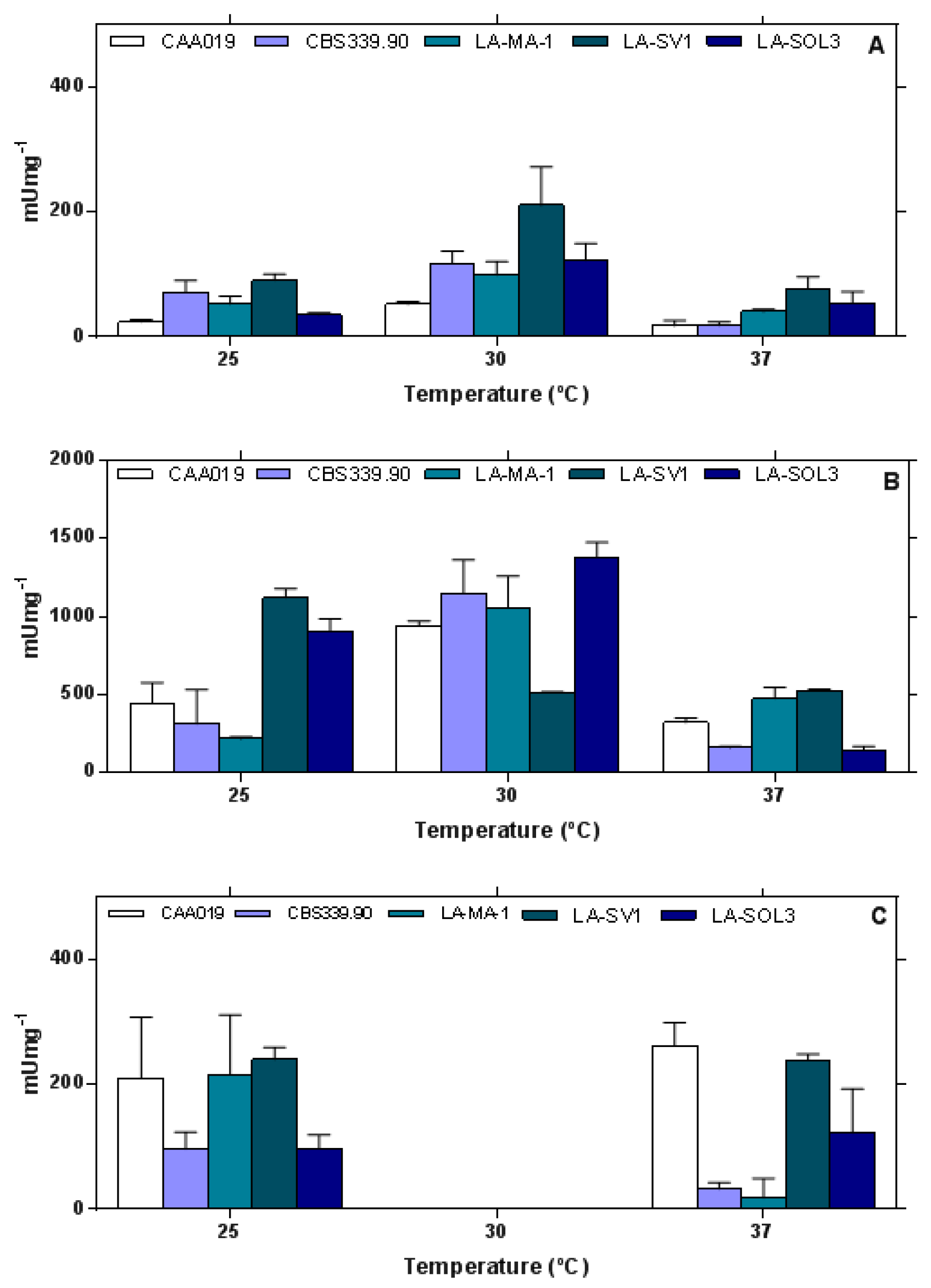
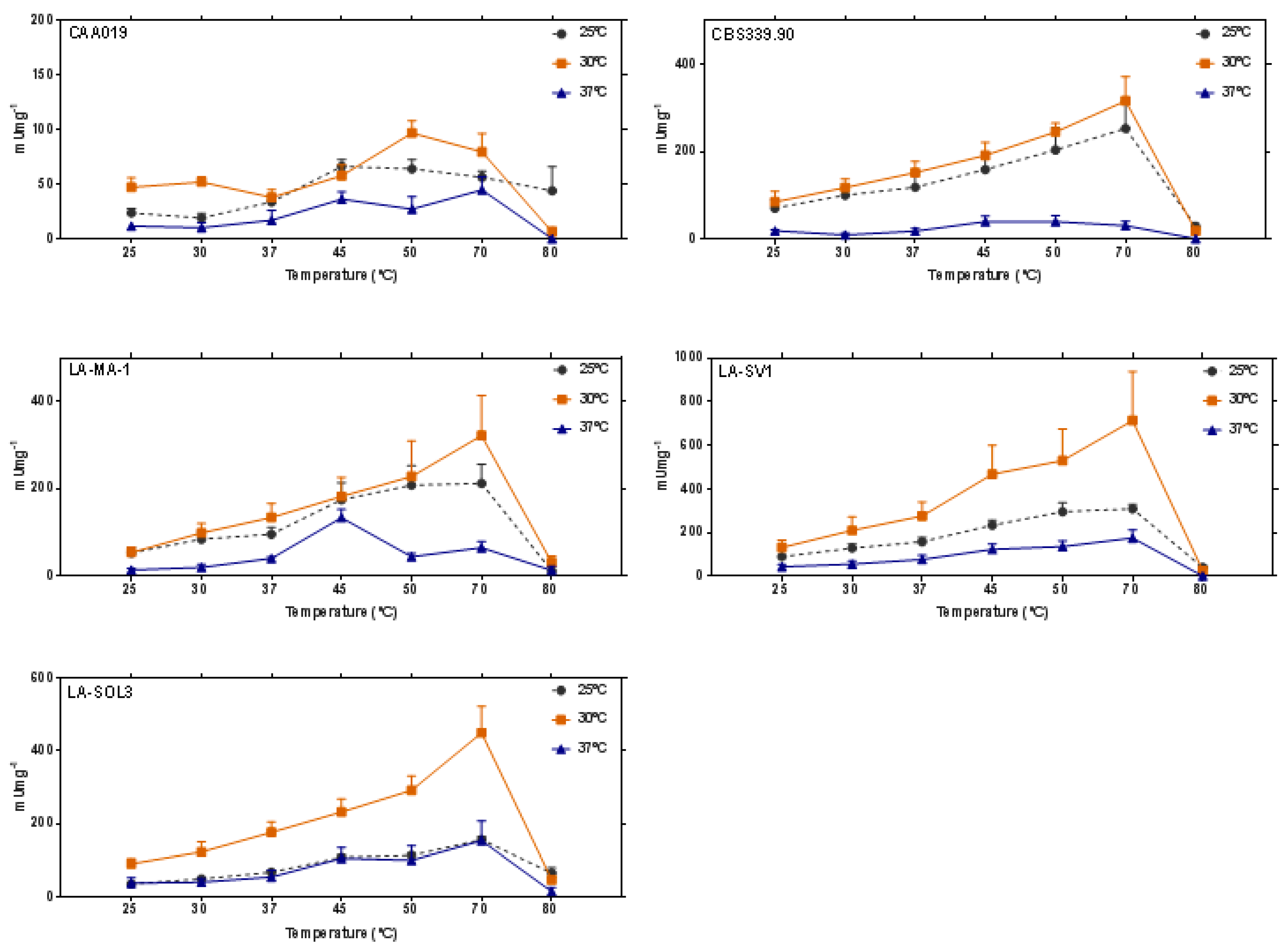
2.2. Thermostability of Endoglucanases
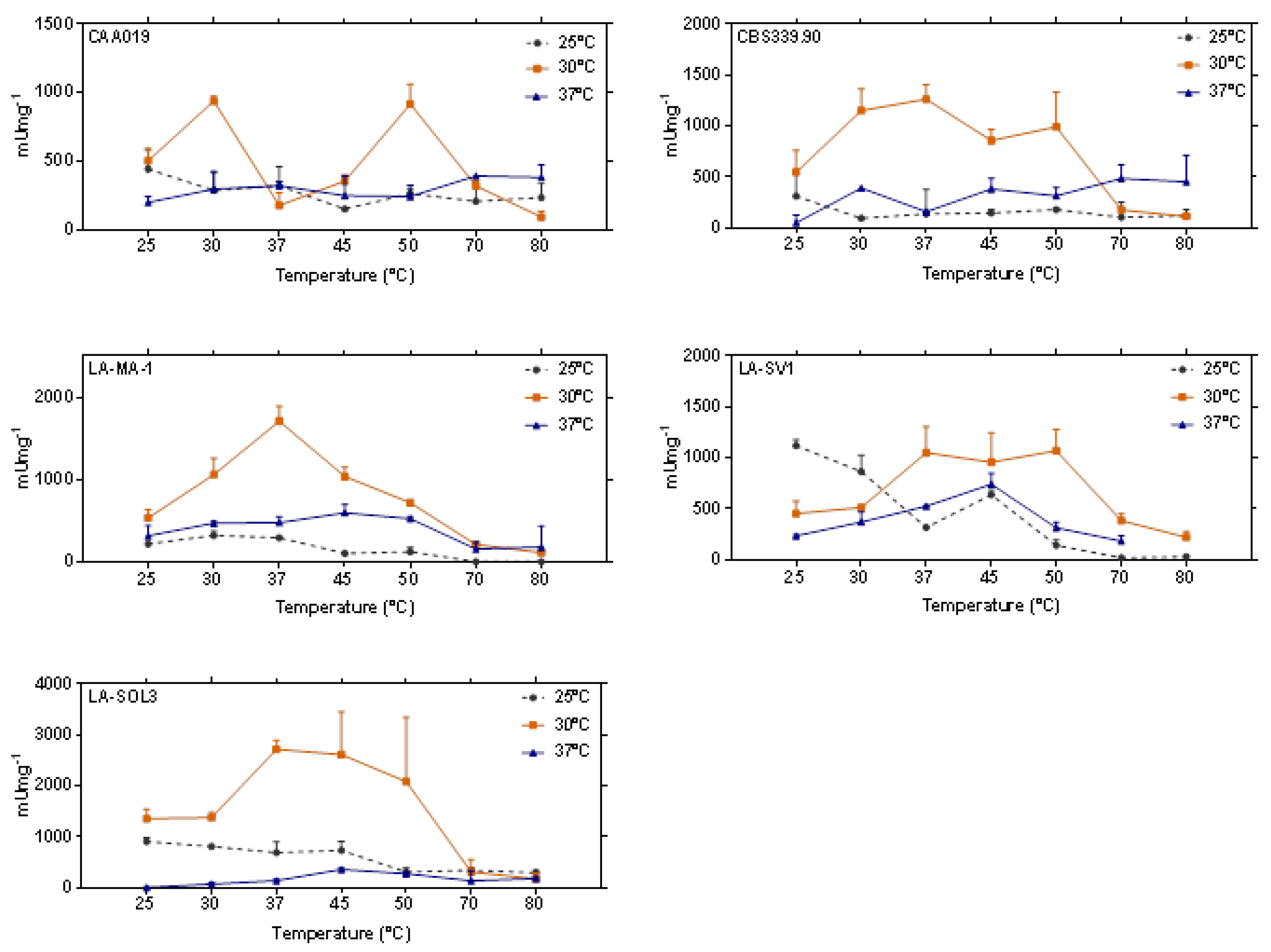
2.3. Thermostability of Proteases
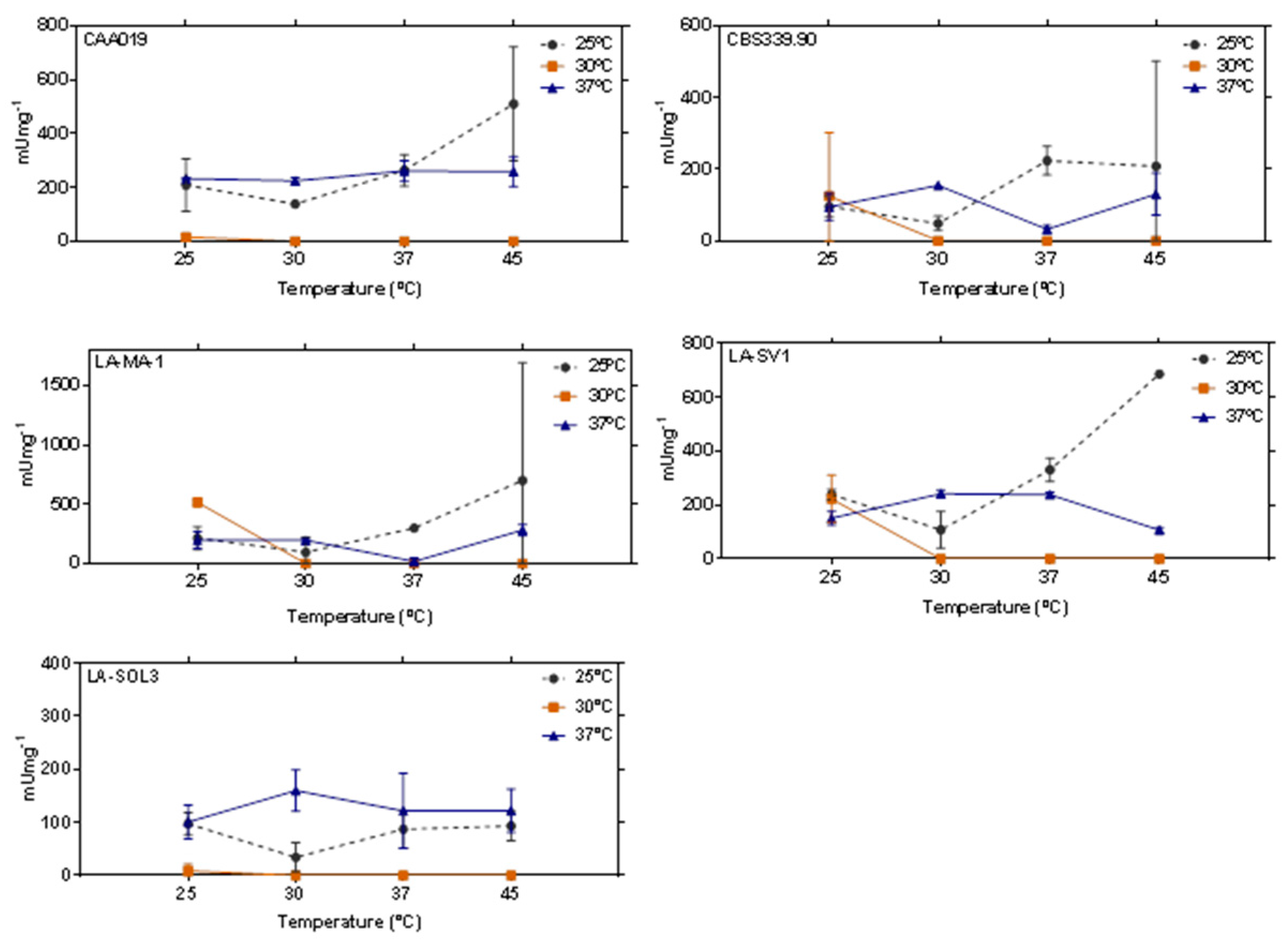
2.4. Thermostability of Lipases
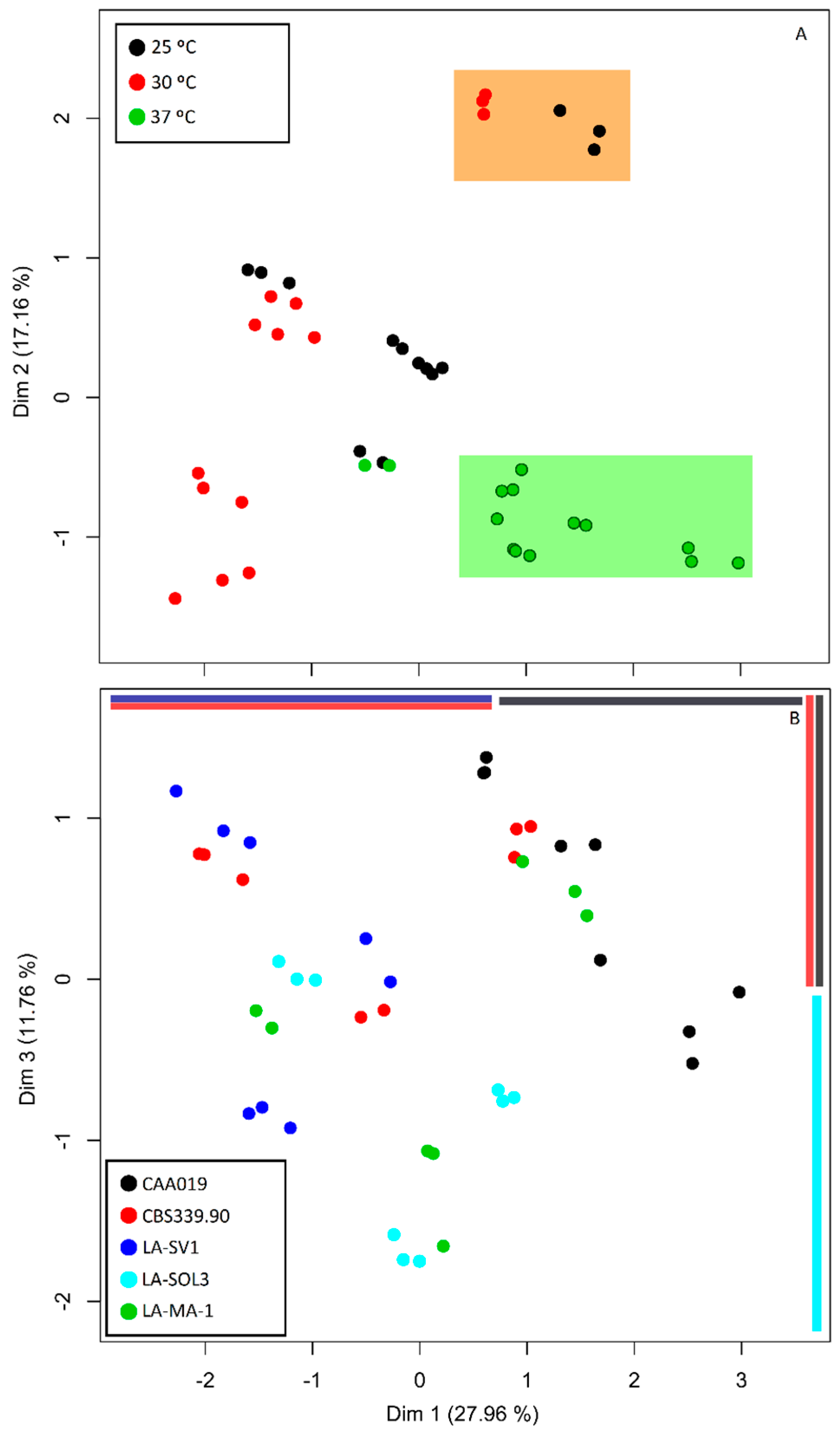
2.5. Multi Factor Analysis of Extracellular Activity
3. Discussion
3.1. Extracellular Enzymatic Activity
3.2. Multi Factor Analysis of Extracellular Activity
3.3. Thermostability
4. Materials and Methods
4.1. Microorganisms
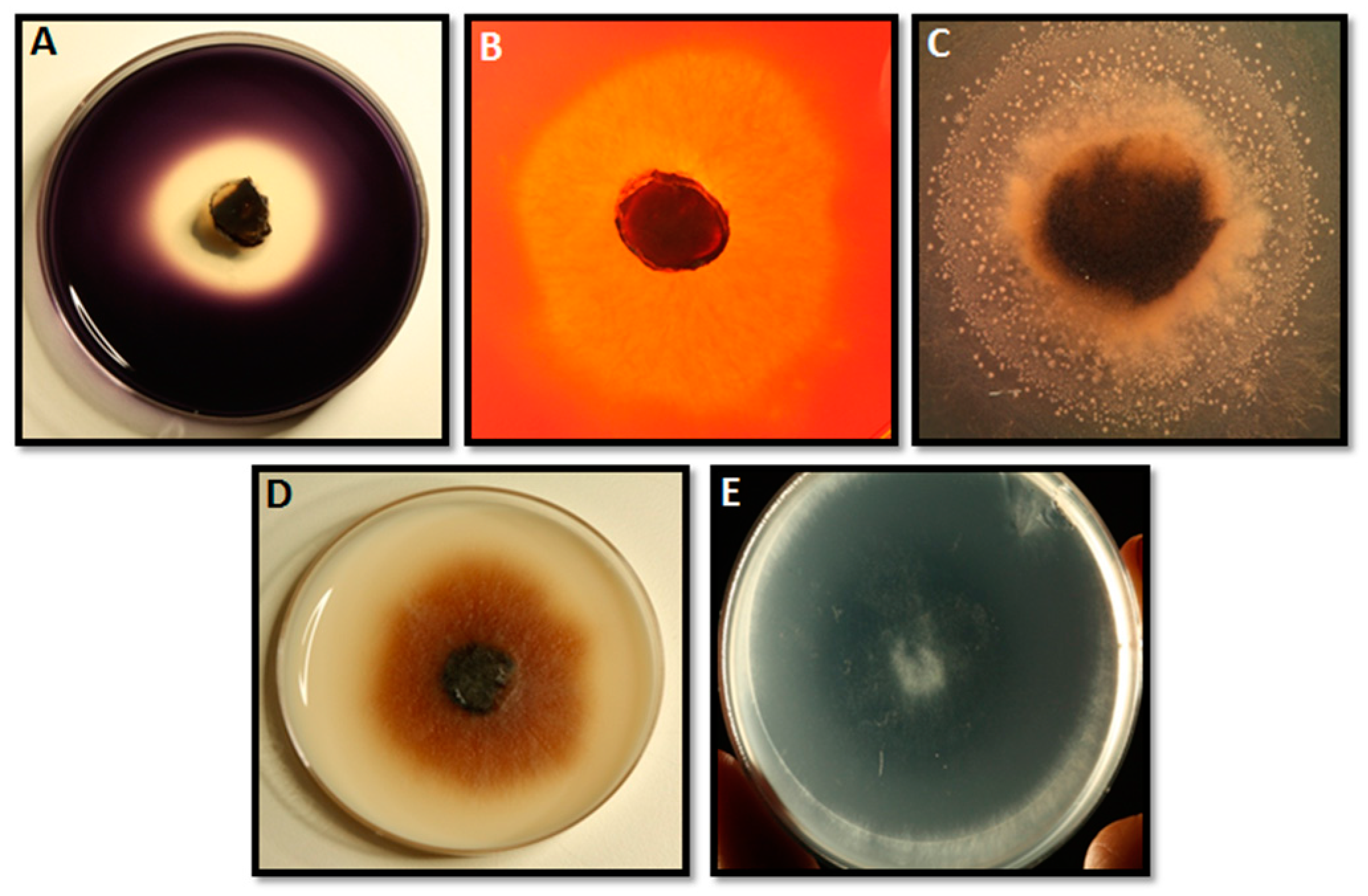
4.2. Detection of Extracellular Enzymes
4.2.1. Proteases
4.2.2. Xylanases and Cellulases
4.2.3. Lipases
4.2.4. Amylases
4.2.5. Pectinases and Pectin Lyases
4.2.6. Laccases
4.3. Quantification of Extracellular Activities
4.3.1. Cellulolytic Activity Quantification
4.3.2. Proteolytic Activity Quantification
4.3.3. Lipolytic Activity Quantification
4.4. Zymography
4.4.1. Cellulases
4.4.2. Proteases
4.4.3. Lipases
4.5. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yike, I. Fungal proteases and their pathophysiological effects. Mycopathologia 2011, 171, 299–323. [Google Scholar] [CrossRef] [PubMed]
- Do Vale, L.H.F.; Gómez-Mendoza, D.P.; Kim, M.S.; Pandey, A.; Ricart, C.A.O.; Edivaldo, X.F.F.; Sousa, M.V. Secretome analysis of the fungus Trichoderma harzianum grown on cellulose. Proteomics 2012, 12, 2716–2728. [Google Scholar] [CrossRef] [PubMed]
- Suryanarayanan, T.S.; Thirunavukkarasu, N.; Govindarajulu, M.B.; Gopalan, V. Fungal endophytes: An untapped source of biocatalysts. Fungal Divers. 2012, 54, 19–30. [Google Scholar] [CrossRef]
- Esteves, A.C.; Saraiva, M.; Correia, A.; Alves, A. Botryosphaeriales fungi produce extracellular enzymes with biotechnological potential. Can. J. Microbiol. 2014, 60, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.M.; King, B.C.; Hayes, M.L.; Bergstrom, G.C. Plant pathogens as a source of diverse enzymes for lignocellulose digestion. Curr. Opin. Microbiol. 2011, 14, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Sajith, S.; Priji, P.; Sreedevi, S.; Benjamin, S. An overview on fungal cellulases with an industrial perspective. J. Nutr. Food Sci. 2016, 6, 461–474. [Google Scholar]
- Park, M.; Do, E.; Jung, W.H. Lipolytic enzymes involved in the virulence of human pathogenic fungi. Mycobiology 2013, 41, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.; Crous, P.W.; Correia, A.; Phillips, A.J.L. Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Divers. 2008, 28, 1–13. [Google Scholar]
- Phillips, A.J.; Alves, A.; Abdollahzadeh, J.; Slippers, B.; Wingfield, M.J.; Groenewald, J.Z.; Crous, P.W. The botryosphaeriaceae: Genera and species known from culture. Stud. Mycol. 2013, 76, 51–167. [Google Scholar] [CrossRef] [PubMed]
- D’souza, A.D.; Maheshwari, R. Senescence in fungi. Resonance 2002, 7, 51–55. [Google Scholar] [CrossRef]
- Jami, F.; Slippers, B.; Wingfield, M.J.; Gryzenhout, M. Greater botryosphaeriaceae diversity in healthy than associated diseased Acacia karroo tree tissues. Austral. Plant Pathol. 2013, 42, 421–430. [Google Scholar] [CrossRef]
- Rodríguez-Gálvez, E.; Maldonado, E.; Alves, A. Identification and pathogenicity of Lasiodiplodia theobromae causing dieback of table grapes in Peru. Eur. J. Plant Pathol. 2015, 141, 477–489. [Google Scholar] [CrossRef]
- Summerbell, R.C.; Krajden, S.; Levine, R.; Fuksa, M. Subcutaneous phaeohyphomycosis caused by Lasiodiplodia theobromae and successfully treated surgically. Med. Mycol. 2004, 42, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Kindo, A.J.; Pramod, C.; Anita, S.; Mohanty, S. Maxillary sinusitis caused by Lasiodiplodia theobromae. Indian J. Med. Microbiol. 2010, 28, 167–169. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Sengupta, J.; Banerjee, D.; Khetan, A. Lasiodiplodia theobromae keratitis: A case report and review of literature. Mycopathologia 2012, 174, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Sengupta, J.; Banerjee, D.; Khetan, A. Lasiodiplodia theobromae keratitis: A rare fungi from eastern india. Microbiology 2013, 3, e19. [Google Scholar] [CrossRef]
- Bennett, J.W. Mycotechnology: The role of fungi in biotechnology. J. Biotechnol. 1998, 66, 101–107. [Google Scholar] [CrossRef]
- Corrêa, R.C.G.; Rhoden, S.A.; Mota, T.R.; Azevedo, J.L.; Pamphile, J.A.; de Souza, C.G.M.; Polizeli, M.L.T.M.; Bracht, A.; Peralta, R.M. Endophytic fungi: Expanding the arsenal of industrial enzyme producers. J. Ind. Microbiol. Biotechnol. 2014, 41, 1467–1478. [Google Scholar] [CrossRef] [PubMed]
- Penttila, M.; Limon, C.; Nevalainen, H. Molecular biology of Trichoderma and biotechnological applications. In Handbook of Fungal Biotechnology; Arora, D.K., Ed.; Marcel Dekker: New York, NY, USA, 2003; pp. 413–427. [Google Scholar]
- Banerjee, A.C.; Kundu, A.; Ghosh, S.K. Genetic manipulation of filamentous fungi. In New Horizons in Biotechnology; Roussos, S., Soccol, C.R., Pandey, A., Augur, C., Eds.; Springer: Dordrecht, The Netherlands, 2003; pp. 193–198. [Google Scholar]
- Blackwell, M. The fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot. 2011, 98, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Braaksma, M.; Punt, P. Aspergillus as a cell factory for protein production: Controlling protease activity in fungal production. In The Aspergilli: Genomics, Medical Aspects, Biotechnology, and Research Methods; Goldman, G.H., Osmani, S.A., Eds.; CRC Press: Boca Raton, FL, USA, 2008; pp. 441–455. [Google Scholar]
- Barros, R.R.O.; Oliveira, R.A.; Gottschalk, L.M.F.; Bon, E.P.S. Production of cellulolytic enzymes by fungi Acrophialophora nainiana and Ceratocystis paradoxa using different carbon sources. Appl. Biochem. Biotechnol. 2010, 161, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Félix, C.; Duarte, A.S.; Vitorino, R.; Guerreiro, A.C.L.; Domingues, P.; Correia, A.C.M.; Alves, A.; Esteves, A.C. Temperature modulates the secretome of the phytopathogenic fungus Lasiodiplodia theobromae. Front. Plant Sci. 2016, 7, 1096. [Google Scholar] [CrossRef] [PubMed]
- King, B.C.; Waxman, K.D.; Nenni, N.V.; Walker, L.P.; Bergstrom, G.C.; Gibson, D.M. Arsenal of plant cell wall degrading enzymes reflects host preference among plant pathogenic fungi. Biotechnol. Biofuels 2011, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Van der Does, H.C.; Rep, M. Virulence genes and the evolution of host specificity in plant-pathogenic fungi. Mol. Plant Microbe Interact. 2007, 20, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Gálvez, E.; Guerrero, P.; Barradas, C.; Crous, P.W.; Alves, A. Phylogeny and pathogenicity of Lasiodiplodia species associated with dieback of mango in Peru. Fungal Biol. 2017, 121, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.A.; Ahmad, I.; Aqil, F.; Owais, M.; Shahid, M.; Musarrat, J. Virulence and pathogenicity of fungal pathogens with special reference to Candida albicans. In Combating Fungal Infections: Problems and Remedy; Ahmad, I., Owais, M., Shahid, M., Aqil, F., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2010; pp. 21–45. [Google Scholar]
- Toth, A.; Nemeth, T.; Csonka, K.; Horvath, P.; Vagvolgyi, C.; Vizler, C.; Nosanchuk, J.D.; Gacser, A. Secreted Candida parapsilosis lipase modulates the immune response of primary human macrophages. Virulence 2014, 5, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.-L. Importance of Secreted Lipases for Virulence of the Phytopathogenic Fungus Fusarium graminearum; University of Hamburg: Hamburg, Germany, 2008. [Google Scholar]
- Reis, H.; Pfiffi, S.; Hahn, M. Molecular and functional characterization of a secreted lipase from Botrytis cinerea. Mol. Plant Pathol. 2005, 6, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Voigt, C.A.; Schafer, W.; Salomon, S. A secreted lipase of Fusarium graminearum is a virulence factor required for infection of cereals. Plant J. 2005, 42, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Abdi, H. Multivariate analysis. In Encyclopedia of Social Sciences Research Methods; Lewis-Beck, M., Bryman, A., Futing, T., Eds.; Sage: Thousand Oaks, CA, USA, 2003. [Google Scholar]
- Duarte, A.S.; Correia, A.; Esteves, A.C. Bacterial collagenases—A review. Crit. Rev. Microbiol. 2016, 42, 106–126. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, M.; Sathiyabama, M. Production, partial purification and characterization of protease from a phytopathogenic fungi Alternaria solani (ell. And mart.) sorauer. J. Basic Microbiol. 2014, 54, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Facchini, F.D.A.; Vici, A.C.; Pereira, M.G.; Jorge, J.A.; Polizeli, M.d.L.T.M. Enhanced lipase production of Fusarium verticillioides by using response surface methodology and wastewater pretreatment application. J. Biochem. Technol. 2015, 6, 996–1002. [Google Scholar]
- Sadati, R.; Barghi, A.; Abbasi Larki, R. Isolation and screening of lipolytic fungi from coastal waters of the southern caspian sea (north of iran). Jundishapur J. Microbiol. 2015, 8, e16426. [Google Scholar] [CrossRef]
- Salihu, A.; Alam, M.Z. Thermostable lipases: An overview of production, purification and characterization. Biosci. Biotechnol. Res. Asia 2014, 11, 1095–1107. [Google Scholar] [CrossRef]
- St Leger, R.J.; Joshi, L.; Roberts, D.W. Adaptation of proteases and carbohydrases of saprophytic, phytopathogenic and entomopathogenic fungi to the requirements of their ecological niches. Microbiology 1997, 143, 1983–1992. [Google Scholar] [CrossRef] [PubMed]
- Hankin, L.; Anagnostakis, S.L. Use of solid media for detection of enzyme production by fungi. Mycologia 1975, 67, 597–607. [Google Scholar] [CrossRef]
- Rigling, D. Isolation and characterization of Cryphonectria parasitica mutants that mimic a specific effect of hypovirulence-associated dsrna on laccase activity. Can. J. Bot. 1995, 73, 1655–1661. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Macchione, M.M.; Merheb, C.W.; Gomes, E.; da Silva, R. Protease production by different thermophilic fungi. Appl. Biochem. Biotechnol. 2008, 146, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Kotogán, A.; Németh, B.; Vágvölgyi, C.; Papp, T.; Takó, M. Screening for extracellular lipase enzymes with transesterification capacity in Mucoromycotina strains. Food Technol. Biotechnol. 2014, 52, 73–82. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage t4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Peterson, R.; Grinyer, J.; Nevalainen, H. Extracellular hydrolase profiles of fungi isolated from koala faeces invite biotechnological interest. Mycol. Prog. 2011, 10, 207–218. [Google Scholar] [CrossRef]
- Duarte, A.S.; Duarte, E.P.; Correia, A.; Pires, E.; Barros, M.T. Cardosins improve neuronal regeneration after cell disruption: A comparative expression study. Cell Biol. Toxicol. 2009, 25, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Castro-Ochoa, L.D.; Rodríguez-Gómez, C.; Valerio-Alfaro, G.; Oliart Ros, R. Screening, purification and characterization of the thermoalkalophilic lipase produced by Bacillus thermoleovorans ccr11. Enzyme Microb. Technol. 2005, 37, 648–654. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2011. [Google Scholar]
- Chavent, M.; Kuentz, V.; Labenne, A.; Liquet, B.; Saracco, J. Pcamixdata: Multivariate Analysis of Mixed Data, R package version 3.0; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Pohlert, T. The Pairwise Multiple Comparison of Mean Ranks Package (PMCMR); R package; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Félix, C.; Libório, S.; Nunes, M.; Félix, R.; Duarte, A.S.; Alves, A.; Esteves, A.C. Lasiodiplodia theobromae as a Producer of Biotechnologically Relevant Enzymes. Int. J. Mol. Sci. 2018, 19, 29. https://doi.org/10.3390/ijms19020029
Félix C, Libório S, Nunes M, Félix R, Duarte AS, Alves A, Esteves AC. Lasiodiplodia theobromae as a Producer of Biotechnologically Relevant Enzymes. International Journal of Molecular Sciences. 2018; 19(2):29. https://doi.org/10.3390/ijms19020029
Chicago/Turabian StyleFélix, Carina, Sofia Libório, Mariana Nunes, Rafael Félix, Ana S. Duarte, Artur Alves, and Ana C. Esteves. 2018. "Lasiodiplodia theobromae as a Producer of Biotechnologically Relevant Enzymes" International Journal of Molecular Sciences 19, no. 2: 29. https://doi.org/10.3390/ijms19020029
APA StyleFélix, C., Libório, S., Nunes, M., Félix, R., Duarte, A. S., Alves, A., & Esteves, A. C. (2018). Lasiodiplodia theobromae as a Producer of Biotechnologically Relevant Enzymes. International Journal of Molecular Sciences, 19(2), 29. https://doi.org/10.3390/ijms19020029