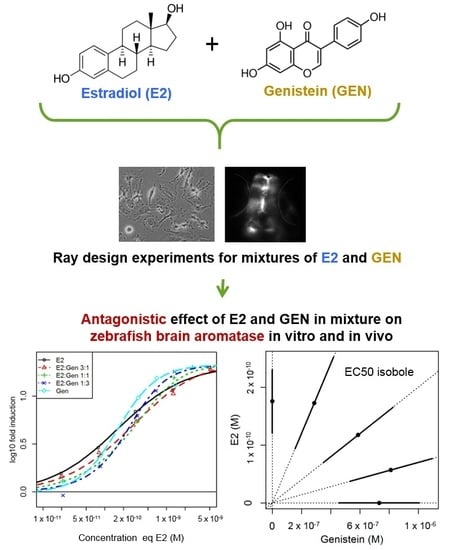
Mixture Concentration-Response Modeling Reveals Antagonistic Effects of Estradiol and Genistein in Combination on Brain Aromatase Gene (cyp19a1b) in Zebrafish
Abstract
:1. Introduction
2. Results
2.1. In Vitro Effects of Single Test Compounds
2.2. In Vitro Effects of Binary Mixtures of E2 and GEN
2.3. In Vivo Effects of Single Test Compounds
2.4. In Vivo Effects of Binary Mixtures of E2 and GEN
3. Discussion
4. Materials and Methods
4.1. Compounds
4.2. Zebrafish Maintenance and Breeding
4.3. Zebrafish Exposure to Estrogenic Compounds
4.4. In Vivo Imaging
4.5. U251-MG Cell Bioassay
4.6. Data Normalization
4.7. Concentration-Response Modeling
4.8. Mixture Experimental Designs
4.9. Mixture Concentration-Response Modeling
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CA | Concentration Addition |
| DL | Dose-Level dependent interaction model |
| E2 | 17β-estradiol |
| EC50 | Effective Concentration 50% |
| ER | Estrogen Receptor |
| FCS | Fetal Calf Serum |
| GEN | Genistein |
| GFP | Green Fluorescent Protein |
| SA | Simple Interaction model |
References
- Jobling, S.; Beresford, N.; Nolan, M.; Rodgers-Gray, T.; Brighty, G.C.; Sumpter, J.P.; Tyler, C.R. Altered sexual maturation and gamete production in wild roach (Rutilus rutilus) living in rivers that receive treated sewage effluents. Biol. Reprod. 2002, 66, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Nash, J.P.; Kime, D.E.; Van der Ven, L.T.; Wester, P.W.; Brion, F.; Maack, G.; Stahlschmidt-Allner, P.; Tyler, C.R. Long-term exposure to environmental concentrations of the pharmaceutical ethynylestradiol causes reproductive failure in fish. Environ. Health Perspect. 2004, 112, 1725–1733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brion, F.; Le Page, Y.; Piccini, B.; Cardoso, O.; Tong, S.K.; Chung, B.C.; Kah, O. Screening estrogenic activities of chemicals or mixtures in vivo using transgenic (cyp19a1b-GFP) zebrafish embryos. PLoS ONE 2012, 7, e36069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kortenkamp, A. Ten years of mixing cocktails: A review of combination effects of endocrine-disrupting chemicals. Environ. Health Perspect. 2007, 115 (Suppl. 1), 98–105. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.L.; Janz, D.M. Effects of binary mixtures of xenoestrogens on gonadal development and reproduction in zebrafish. Aquat. Toxicol. 2006, 80, 382–395. [Google Scholar] [CrossRef] [PubMed]
- Petersen, K.; Fetter, E.; Kah, O.; Brion, F.; Scholz, S.; Tollefsen, K.E. Transgenic (cyp19a1b-GFP) zebrafish embryos as a tool for assessing combined effects of oestrogenic chemicals. Aquat. Toxicol. 2013, 138–139, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Petersen, K.; Tollefsen, K.E. Assessing combined toxicity of estrogen receptor agonists in a primary culture of rainbow trout (Oncorhynchus mykiss) hepatocytes. Aquat. Toxicol. 2011, 101, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Rajapakse, N.; Silva, E.; Scholze, M.; Kortenkamp, A. Deviation from additivity with estrogenic mixtures containing 4-nonylphenol and 4-tert-octylphenol detected in the e-screen assay. Environ. Sci. Technol. 2004, 38, 6343–6352. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, K.L.; Hutchinson, T.H.; Hetheridge, M.J.; Scholze, M.; Sumpter, J.P.; Tyler, C.R. Assessing the biological potency of binary mixtures of environmental estrogens using vitellogenin induction in juvenile rainbow trout (Oncorhynchus mykiss). Environ. Sci. Technol. 2001, 35, 2476–2481. [Google Scholar] [CrossRef] [PubMed]
- Garriz, A.; Menendez-Helman, R.J.; Miranda, L.A. Effects of estradiol and ethinylestradiol on sperm quality, fertilization, and embryo-larval survival of pejerrey fish (Odontesthes bonariensis). Aquat. Toxicol. 2015, 167, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Song, W.T.; Wang, Z.J.; Liu, H.C. Effects of individual and binary mixtures of estrogens on male goldfish (Carassius auratus). Fish Physiol. Biochem. 2014, 40, 1927–1935. [Google Scholar] [CrossRef] [PubMed]
- Hinfray, N.; Tebby, C.; Garoche, C.; Piccini, B.; Bourgine, G.; Ait-Aissa, S.; Kah, O.; Pakdel, F.; Brion, F. Additive effects of levonorgestrel and ethinylestradiol on brain aromatase (cyp19a1b) in zebrafish specific in vitro and in vivo bioassays. Toxicol. Appl. Pharmacol. 2016, 307, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Diotel, N.; Vaillant, C.; Gabbero, C.; Mironov, S.; Fostier, A.; Gueguen, M.-M.; Anglade, I.; Kah, O.; Pellegrini, E. Effects of estradiol in adult neurogenesis and brain repair in zebrafish. Horm. Behav. 2013, 63, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, E.; Mouriec, K.; Anglade, I.; Menuet, A.; Le Page, Y.; Gueguen, M.M.; Marmignon, M.H.; Brion, F.; Pakdel, F.; Kah, O. Identification of aromatase-positive radial glial cells as progenitor cells in the ventricular layer of the forebrain in zebrafish. J. Comp. Neurol. 2007, 501, 150–167. [Google Scholar] [CrossRef] [PubMed]
- Le Page, Y.; Menuet, A.; Kah, O.; Pakdel, F. Characterization of a cis-acting element involved in cell-specific expression of the zebrafish brain aromatase gene. Mol. Reprod. Dev. 2008, 75, 1549–1557. [Google Scholar] [CrossRef] [PubMed]
- Le Page, Y.; Scholze, M.; Kah, O.; Pakdel, F. Assessment of xenoestrogens using three distinct estrogen receptors and the zebrafish brain aromatase gene in a highly responsive glial cell system. Environ. Health Perspect. 2006, 114, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Lecomte, S.; Demay, F.; Ferriere, F.; Pakdel, F. Phytochemicals targeting estrogen receptors: Beneficial rather than adverse effects? Int. J. Mol. Sci. 2017, 18, 1381. [Google Scholar] [CrossRef]
- Jonker, M.J.; Svendsen, C.; Bedaux, J.J.; Bongers, M.; Kammenga, J.E. Significance testing of synergistic/antagonistic, dose level-dependent, or dose ratio-dependent effects in mixture dose-response analysis. Environ. Toxicol. Chem. SETAC 2005, 24, 2701–2713. [Google Scholar] [CrossRef]
- Loewe, S. The problem of synergism and antagonism of combined drugs. Arzneimittelforschung 1953, 3, 285–290. [Google Scholar] [PubMed]
- Legler, J.; Zeinstra, L.M.; Schuitemaker, F.; Lanser, P.H.; Bogerd, J.; Brouwer, A.; Vethaak, A.D.; De Voogt, P.; Murk, A.J.; Van der Burg, B. Comparison of in vivo and in vitro reporter gene assays for short-term screening of estrogenic activity. Environ. Sci. Technol. 2002, 36, 4410–4415. [Google Scholar] [CrossRef] [PubMed]
- Cosnefroy, A.; Brion, F.; Maillot-Marechal, E.; Porcher, J.M.; Pakdel, F.; Balaguer, P.; Ait-Aissa, S. Selective activation of zebrafish estrogen receptor subtypes by chemicals by using stable reporter gene assay developed in a zebrafish liver cell line. Toxicol. Sci. 2012, 125, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Jarošová, B.; Javůrek, J.; Adamovský, O.; Hilscherová, K. Phytoestrogens and mycoestrogens in surface waters—Their sources, occurrence, and potential contribution to estrogenic activity. Environ. Int. 2015, 81, 26–44. [Google Scholar] [CrossRef] [PubMed]
- Escande, A.; Pillon, A.; Servant, N.; Cravedi, J.-P.; Larrea, F.; Muhn, P.; Nicolas, J.-C.; Cavaillès, V.; Balaguer, P. Evaluation of ligand selectivity using reporter cell lines stably expressing estrogen receptor alpha or beta. Biochem. Pharmacol. 2006, 71, 1459–1469. [Google Scholar] [CrossRef] [PubMed]
- Pillon, A.; Servant, N.; Vignon, F.; Balaguer, P.; Nicolas, J.C. In vivo bioluminescence imaging to evaluate estrogenic activities of endocrine disrupters. Anal. Biochem. 2005, 340, 295–302. [Google Scholar] [CrossRef]
- Cosnefroy, A.; Brion, F.; Guillet, B.; Laville, N.; Porcher, J.M.; Balaguer, P.; Ait-Aissa, S. A stable fish reporter cell line to study estrogen receptor transactivation by environmental (xeno)estrogens. Toxicol. In Vitro 2009, 23, 1450–1454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiller, V.; Wichmann, A.; Kriehuber, R.; Muth-Kohne, E.; Giesy, J.P.; Hecker, M.; Fenske, M. Studying the effects of genistein on gene expression of fish embryos as an alternative testing approach for endocrine disruption. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2013, 157, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Soto, A.M.; Fernandez, M.F.; Luizzi, M.F.; Oles Karasko, A.S.; Sonnenschein, C. Developing a marker of exposure to xenoestrogen mixtures in human serum. Environ. Health Perspect. 1997, 105 (Suppl. 3), 647–654. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Ide, K.; Ishida, M. Response of mcf-7 human breast cancer cells to some binary mixtures of oestrogenic compounds in vitro. J. Pharm. Pharmacol. 2001, 53, 1549–1554. [Google Scholar] [CrossRef] [PubMed]
- Kunz, P.Y.; Fent, K. Estrogenic activity of ternary UV filter mixtures in fish (Pimephales promelas)—An analysis with nonlinear isobolograms. Toxicol. Appl. Pharmacol. 2009, 234, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Van Meeuwen, J.A.; ter Burg, W.; Piersma, A.H.; van den Berg, M.; Sanderson, J.T. Mixture effects of estrogenic compounds on proliferation and pS2 expression of MCF-7 human breast cancer cells. Food Chem. Toxicol. 2007, 45, 2319–2330. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Edwards, R.J.; Boobis, A.R. Proteomic analysis of human breast cell lines using SELDI-TOF MS shows that mixtures of estrogenic compounds exhibit simple similar action (concentration additivity). Toxicol. Lett. 2008, 181, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Charles, G.D.; Gennings, C.; Zacharewski, T.R.; Gollapudi, B.B.; Carney, E.W. Assessment of interactions of diverse ternary mixtures in an estrogen receptor-α reporter assay. Toxicol. Appl. Pharmacol. 2002, 180, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Routledge, E.J.; White, R.; Parker, M.G.; Sumpter, J.P. Differential effects of xenoestrogens on coactivator recruitment by estrogen receptor (ER) α and ERβ. J. Biol. Chem. 2000, 275, 35986–35993. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.C.; Charn, T.H.; Park, S.-H.; Helferich, W.G.; Komm, B.; Katzenellenbogen, J.A.; Katzenellenbogen, B.S. Estrogen receptors α and β as determinants of gene expression: Influence of ligand, dose, and chromatin binding. Mol. Endocrinol. 2008, 22, 1032–1043. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.K.; Mouriec, K.; Kuo, M.W.; Pellegrini, E.; Gueguen, M.M.; Brion, F.; Kah, O.; Chung, B.C. A cyp19a1b-GFP (aromatase B) transgenic zebrafish line that expresses GFP in radial glial cells. Genesis 2009, 47, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Menuet, A.; Pellegrini, E.; Anglade, I.; Blaise, O.; Laudet, V.; Kah, O.; Pakdel, F. Molecular characterization of three estrogen receptor forms in zebrafish: Binding characteristics, transactivation properties, and tissue distributions. Biol. Reprod. 2002, 66, 1881–1892. [Google Scholar] [CrossRef] [PubMed]
- Menuet, A.; Pellegrini, E.; Brion, F.; Gueguen, M.M.; Anglade, I.; Pakdel, F.; Kah, O. Expression and estrogen-dependent regulation of the zebrafish brain aromatase gene. J. Comp. Neurol. 2005, 485, 304–320. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Ritz, C.; Streibig, J.C. Bioassays analysis using R. J. Stat. Softw. 2005, 12, 1–22. [Google Scholar] [CrossRef]
- Berenbaum, M.C. The expected effect of a combination of agents: The general solution. J. Theor. Biol. 1985, 114, 413–431. [Google Scholar] [CrossRef]
- Gennings, C.; Carter, W.H., Jr.; Carchman, R.A.; Teuschler, L.K.; Simmons, J.E.; Carney, E.W. A unifying concept for assessing toxicological interactions: Changes in slope. Toxicol. Sci. 2005, 88, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Backhaus, T.; Arrhenius, A.; Blanck, H. Toxicity of a mixture of dissimilarly acting substances to natural algal communities: Predictive power and limitations of independent action and concentration addition. Environ. Sci. Technol. 2004, 38, 6363–6370. [Google Scholar] [CrossRef] [PubMed]
- Webster, T.F. Mixtures of endocrine disruptors: How similar must mechanisms be for concentration addition to apply? Toxicology 2013, 313, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Faust, M.; Altenburger, R.; Backhaus, T.; Blanck, H.; Boedeker, W.; Gramatica, P.; Hamer, V.; Scholze, M.; Vighi, M.; Grimme, L.H. Predicting the joint algal toxicity of multi-component s-triazine mixtures at low-effect concentrations of individual toxicants. Aquat. Toxicol. 2001, 56, 13–32. [Google Scholar] [CrossRef]
- Goldoni, M.; Johansson, C. A mathematical approach to study combined effects of toxicants in vitro: Evaluation of the bliss independence criterion and the loewe additivity model. Toxicol. In Vitro 2007, 21, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Thienpont, B.; Barata, C.; Raldùa, D. Modeling mixtures of thyroid gland function disruptors in a vertebrate alternative model, the zebrafish eleutheroembryo. Toxicol. Appl. Pharmacol. 2013, 269, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Greco, W.R.; Bravo, G.; Parsons, J.C. The search for synergy—A critical-review from a response-surface perspective. Pharmacol. Rev. 1995, 47, 331–385. [Google Scholar] [PubMed]
- Gessner, P.K. Isobolographic analysis of interactions: An update on applications and utility. Toxicology 1995, 105, 161–179. [Google Scholar] [CrossRef]
- Kortenkamp, A.; Altenburger, R. Synergisms with mixtures of xenoestrogens: A reevaluation using the method of isoboles. Sci. Total Environ. 1998, 221, 59–73. [Google Scholar] [CrossRef]
- Chou, T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.C.; Talalay, P. Quantitative-analysis of dose-effect relationships—The combined effects of multiple-drugs or enzyme-inhibitors. Adv. Enzym. Regul. 1984, 22, 27–55. [Google Scholar] [CrossRef]
- Gennings, C.; Carter, W.H. Utilizing concentration-response data from individual components to detect statistically significant departures from additivity in chemical mixtures. Biometrics 1995, 51, 1264–1277. [Google Scholar] [CrossRef]
- Boik, J.C.; Newman, R.A.; Boik, R.J. Quantifying synergism/antagonism using nonlinear mixed-effects modeling: A simulation study. Stat. Med. 2008, 27, 1040–1061. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, H.; Cedergreen, N.; Skovgaard, I.; Streibig, J.C. An isobole-based statistical model and test for synergism/antagonism in binary mixture toxicity experiments. Environ. Ecol. Stat. 2007, 14, 383–397. [Google Scholar] [CrossRef]
- Varadhan, R.; Borchers, H.W.; ABB Corporate Research. Dfoptim: Derivative-Free Optimization; Ravi Hopkins University: Baltimore, MD, USA, 2011. [Google Scholar]





| Condition | [E2] (M) | [GEN] (M) | Ray |
|---|---|---|---|
| 1 | 0 | 0 | - |
| 2 | 5.00 × 10−9 | 0 | 1:0 |
| 3 | 1.25 × 10−9 | 0 | 1:0 |
| 4 | 3.12 × 10−10 | 0 | 1:0 |
| 5 | 7.81 × 10−11 | 0 | 1:0 |
| 6 | 1.95 × 10−11 | 0 | 1:0 |
| 7 | 3.75 × 10−9 | 6.25 × 10−6 | 3:1 |
| 8 | 9.37 × 10−10 | 1.56 × 10−6 | 3:1 |
| 9 | 2.34 × 10−10 | 3.91 × 10−7 | 3:1 |
| 10 | 5.86 × 10−11 | 9.77 × 10−8 | 3:1 |
| 11 | 1.46 × 10−11 | 2.44 × 10−8 | 3:1 |
| 12 | 1.25 × 10−9 | 6.25 × 10−6 | 1:1 |
| 13 | 6.25 × 10−10 | 3.12 × 10−6 | 1:1 |
| 14 | 1.56 × 10−10 | 7.81 × 10−7 | 1:1 |
| 15 | 3.91 × 10−11 | 1.95 × 10−7 | 1:1 |
| 16 | 9.77 × 10−12 | 4.88 × 10−8 | 1:1 |
| 17 | 3.12 × 10−10 | 4.69 × 10−6 | 1:3 |
| 18 | 1.56 × 10−10 | 2.34 × 10−6 | 1:3 |
| 19 | 7.81 × 10−11 | 1.17 × 10−6 | 1:3 |
| 20 | 1.95 × 10−11 | 2.93 × 10−7 | 1:3 |
| 21 | 4.88 × 10−12 | 7.32 × 10−8 | 1:3 |
| 22 | 0 | 6.25 × 10−6 | 0:1 |
| 23 | 0 | 3.12 × 10−6 | 0:1 |
| 24 | 0 | 1.56 × 10−6 | 0:1 |
| 25 | 0 | 3.91 × 10−7 | 0:1 |
| 26 | 0 | 9.77 × 10−8 | 0:1 |
| ERα | ERβ2 | ||||
|---|---|---|---|---|---|
| Condition | [E2] (M) | [GEN] (M) | [E2] (M) | [GEN] (M) | Ray |
| 1 | 0 | 0 | 0 | 0 | - |
| 2 | 2.00 × 10−8 | 0 | 2.00 × 10−8 | 0 | 1:0 |
| 3 | 2.00 × 10−9 | 0 | 2.00 × 10−9 | 0 | 1:0 |
| 4 | 2.00 × 10−10 | 0 | 2.00 × 10−10 | 0 | 1:0 |
| 5 | 2.00 × 10−11 | 0 | 2.00 × 10−11 | 0 | 1:0 |
| 6 | 2.00 × 10−12 | 0 | 2.00 × 10−12 | 0 | 1:0 |
| 7 | 1.50 × 10−8 | 1.00 × 10−6 | 1.50 × 10−8 | 2.00 × 10−7 | 3:1 |
| 8 | 1.50 × 10−9 | 1.00 × 10−7 | 1.50 × 10−9 | 2.00 × 10−8 | 3:1 |
| 9 | 1.50 × 10−10 | 1.00 × 10−8 | 1.50 × 10−10 | 2.00 × 10−9 | 3:1 |
| 10 | 1.50 × 10−11 | 1.00 × 10−9 | 1.50 × 10−11 | 2.00 × 10−10 | 3:1 |
| 11 | 1.50 × 10−12 | 1.00 × 10−10 | 1.50 × 10−12 | 2.00 × 10−11 | 3:1 |
| 12 | 1.00 × 10−8 | 2.00 × 10−6 | 1.00 × 10−8 | 4.00 × 10−7 | 1:1 |
| 13 | 1.00 × 10−9 | 2.00 × 10−7 | 1.00 × 10−9 | 4.00 × 10−8 | 1:1 |
| 14 | 1.00 × 10−10 | 2.00 × 10−8 | 1.00 × 10−10 | 4.00 × 10−9 | 1:1 |
| 15 | 1.00 × 10−11 | 2.00 × 10−9 | 1.00 × 10−11 | 4.00 × 10−10 | 1:1 |
| 16 | 1.00 × 10−12 | 2.00 × 10−10 | 1.00 × 10−12 | 4.00 × 10−11 | 1:1 |
| 17 | 5.00 × 10−9 | 3.00 × 10−6 | 5.00 × 10−9 | 6.00 × 10−7 | 1:3 |
| 18 | 5.00 × 10−10 | 3.00 × 10−7 | 5.00 × 10−10 | 6.00 × 10−8 | 1:3 |
| 19 | 5.00 × 10−11 | 3.00 × 10−8 | 5.00 × 10−11 | 6.00 × 10−9 | 1:3 |
| 20 | 5.00 × 10−12 | 3.00 × 10−9 | 5.00 × 10−12 | 6.00 × 10−10 | 1:3 |
| 21 | 5.00 × 10−13 | 3.00 × 10−10 | 5.00 × 10−13 | 6.00 × 10−11 | 1:3 |
| 22 | 0 | 4.00 × 10−6 | 0 | 8.00 × 10−7 | 0:1 |
| 23 | 0 | 4.00 × 10−7 | 0 | 8.00 × 10−8 | 0:1 |
| 24 | 0 | 4.00 × 10−8 | 0 | 8.00 × 10−9 | 0:1 |
| 25 | 0 | 4.00 × 10−9 | 0 | 8.00 × 10−10 | 0:1 |
| 26 | 0 | 4.00 × 10−10 | 0 | 8.00 × 10−11 | 0:1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hinfray, N.; Tebby, C.; Piccini, B.; Bourgine, G.; Aït-Aïssa, S.; Porcher, J.-M.; Pakdel, F.; Brion, F. Mixture Concentration-Response Modeling Reveals Antagonistic Effects of Estradiol and Genistein in Combination on Brain Aromatase Gene (cyp19a1b) in Zebrafish. Int. J. Mol. Sci. 2018, 19, 1047. https://doi.org/10.3390/ijms19041047
Hinfray N, Tebby C, Piccini B, Bourgine G, Aït-Aïssa S, Porcher J-M, Pakdel F, Brion F. Mixture Concentration-Response Modeling Reveals Antagonistic Effects of Estradiol and Genistein in Combination on Brain Aromatase Gene (cyp19a1b) in Zebrafish. International Journal of Molecular Sciences. 2018; 19(4):1047. https://doi.org/10.3390/ijms19041047
Chicago/Turabian StyleHinfray, Nathalie, Cleo Tebby, Benjamin Piccini, Gaelle Bourgine, Sélim Aït-Aïssa, Jean-Marc Porcher, Farzad Pakdel, and François Brion. 2018. "Mixture Concentration-Response Modeling Reveals Antagonistic Effects of Estradiol and Genistein in Combination on Brain Aromatase Gene (cyp19a1b) in Zebrafish" International Journal of Molecular Sciences 19, no. 4: 1047. https://doi.org/10.3390/ijms19041047