Induced Tissue-Specific Stem Cells and Epigenetic Memory in Induced Pluripotent Stem Cells
Abstract
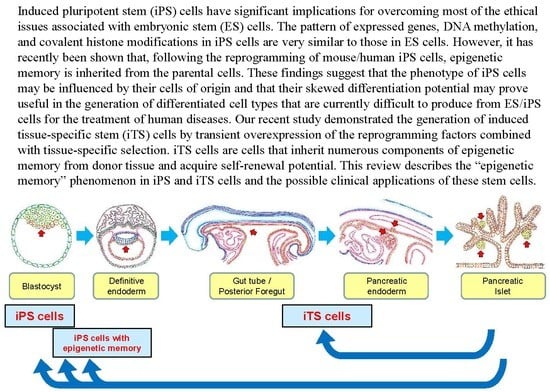
:1. Introduction
2. Generation of iPS Cells
3. Properties of iPS Cells Imbued by Epigenetic Memory
4. DNA Methylation in iPS Cells
5. Tissue-Specific Stem Cells
6. Induced Tissue-Specific Stem Cells
7. Epigenetic Variation between iPS Cell Lines
8. Cancer Development through the Transient Overexpression of Reprogramming Factors
9. Conclusions
Acknowledgements
Author Contributions
Conflicts of Interest
Abbreviations
| iPS | induced pluripotent stem |
| ES | embryonic stem |
| iTS | induced tissue-specific stem |
| iTS-P | iTS cells from the pancreas |
| iTS-L | iTS cells from the liver |
| DMRs | differentially methylated regions |
| Pdx1 | pancreatic and duodenal homeobox factor-1 |
| qRT-PCR | quantitative reverse transcription polymerase chain reaction |
References
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Park, I.H.; Zhao, R.; West, J.A.; Yabuuchi, A.; Huo, H.; Ince, T.A.; Lerou, P.H.; Lensch, M.W.; Daley, G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 2008, 451, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Maherali, N.; Sridharan, R.; Xie, W.; Utikal, J.; Eminli, S.; Arnold, K.; Stadtfeld, M.; Yachechko, R.; Tchieu, J.; Jaenisch, R.; et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 2007, 1, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Ichisaka, T.; Yamanaka, S. Generation of germline-competent induced pluripotent stem cells. Nature 2007, 448, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Wernig, M.; Meissner, A.; Foreman, R.; Brambrink, T.; Ku, M.; Hochedlinger, K.; Bernstein, B.E.; Jaenisch, R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 2007, 448, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Nakagawa, M.; Hyenjong, H.; Ichisaka, T.; Yamanaka, S. Generation of mouse induced pluripotent stem cells without viral vectors. Science 2008, 322, 949–953. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Matsumura, Y.; Sato, Y.; Okada, A.; Morizane, A.; Okamoto, S.; Hong, H.; Nakagawa, M.; Tanabe, K.; Tezuka, K.; et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods 2011, 8, 409–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Hu, K.; Smuga-Otto, K.; Tian, S.; Stewart, R.; Slukvin, I.I.; Thomson, J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science 2009, 324, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Stadtfeld, M.; Nagaya, M.; Utikal, J.; Weir, G.; Hochedlinger, K. Induced pluripotent stem cells generated without viral integration. Science 2008, 322, 945–949. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, N.; Gros, E.; Li, H.R.; Kumar, S.; Deacon, D.C.; Maron, C.; Muotri, A.R.; Chi, N.C.; Fu, X.D.; Yu, B.D.; et al. Efficient generation of human iPSCs by a synthetic self-replicative RNA. Cell Stem Cell 2013, 13, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, H.; Saitoh, I.; Tsugata, T.; Kataoka, H.; Watanabe, M.; Noguchi, Y. Induction of tissue-specific stem cells by reprogramming factors, and tissue-specific selection. Cell Death Differ. 2015, 22, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, I.; Sato, M.; Soda, M.; Inada, E.; Iwase, Y.; Murakami, T.; Ohshima, H.; Hayasaki, H.; Noguchi, H. Tissue-Specific Stem Cells Obtained by Reprogramming of Non-Obese Diabetic (NOD) Mouse-Derived Pancreatic Cells Confer Insulin Production in Response to Glucose. PLoS ONE 2016, 11, e0163580. [Google Scholar] [CrossRef] [PubMed]
- Bar-Nur, O.; Russ, H.A.; Efrat, S.; Benvenisty, N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell 2011, 9, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.K.; Smith, Z.D.; Meissner, A.; et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Chin, M.H.; Mason, M.J.; Xie, W.; Volinia, S.; Singer, M.; Peterson, C.; Ambartsumyan, G.; Aimiuwu, O.; Richter, L.; Zhang, J.; et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 2009, 5, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, Z.; Wilson, K.D.; Wu, Y.; Hu, S.; Quertermous, T.; Wu, J.C. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS ONE 2010, 5, e8975. [Google Scholar] [CrossRef] [PubMed]
- Marchetto, M.C.; Yeo, G.W.; Kainohana, O.; Marsala, M.; Gage, F.H.; Muotri, A.R. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS ONE 2009, 4, e7076. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Lu, S.J.; Klimanskaya, I.; Gomes, I.; Kim, D.; Chung, Y.; Honig, G.R.; Kim, K.S.; Lanza, R. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells 2010, 28, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.Y.; Weick, J.P.; Yu, J.; Ma, L.X.; Zhang, X.Q.; Thomson, J.A.; Zhang, S.C. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA 2010, 107, 4335–4340. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Doi, A.; Wen, B.; Ng, K.; Zhao, R.; Cahan, P.; Kim, J.; Aryee, M.J.; Ji, H.; Ehrlich, L.I.; et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010, 467, 285–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polo, J.M.; Liu, S.; Figueroa, M.E.; Kulalert, W.; Eminli, S.; Tan, K.Y.; Apostolou, E.; Stadtfeld, M.; Li, Y.; Shioda, T.; et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010, 28, 848–855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lister, R.; Pelizzola, M.; Kida, Y.S.; Hawkins, R.D.; Nery, J.R.; Hon, G.; Antosiewicz-Bourget, J.; O’Malley, R.; Castanon, R.; Klugman, S.; et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 2011, 471, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Doi, A.; Park, I.H.; Wen, B.; Murakami, P.; Aryee, M.J.; Irizarry, R.; Herb, B.; Ladd-Acosta, C.; Rho, J.; Loewer, S.; et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009, 41, 1350–1353. [Google Scholar] [CrossRef] [PubMed]
- Ohi, Y.; Qin, H.; Hong, C.; Blouin, L.; Polo, J.M.; Guo, T.; Qi, Z.; Downey, S.L.; Manos, P.D.; Rossi, D.J.; et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell Biol. 2011, 13, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Zhao, R.; Doi, A.; Ng, K.; Unternaehrer, J.; Cahan, P.; Huo, H.; Loh, Y.H.; Aryee, M.J.; Lensch, M.W.; et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat. Biotechnol. 2011, 29, 1117–1119. [Google Scholar] [CrossRef] [PubMed]
- Stadtfeld, M.; Apostolou, E.; Akutsu, H.; Fukuda, A.; Follett, P.; Natesan, S.; Kono, T.; Shioda, T.; Hochedlinger, K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 2010, 465, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Toyoda, M.; Yamazaki-Inoue, M.; Fukawatase, Y.; Chikazawa, E.; Sakaguchi, H.; Akutsu, H.; Umezawa, A. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 2011, 7, e1002085. [Google Scholar] [CrossRef] [PubMed]
- Kaminsky, Z.A.; Tang, T.; Wang, S.C.; Ptak, C.; Oh, G.H.; Wong, A.H.; Feldcamp, L.A.; Virtanen, C.; Halfvarson, J.; Tysk, C.; et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat. Genet. 2009, 41, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Shao, K.; Koch, C.; Gupta, M.K.; Lin, Q.; Lenz, M.; Laufs, S.; Denecke, B.; Schmidt, M.; Linke, M.; Hennies, H.C.; et al. Induced pluripotent mesenchymal stromal cell clones retain donor-derived differences in DNA methylation profiles. Mol. Ther. 2013, 21, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Weissman, I.L. Translating stem and progenitor cell biology to the clinic: Barriers and opportunities. Science 2000, 287, 1442–1446. [Google Scholar] [CrossRef] [PubMed]
- Potten, C.S.; Grant, H.K. The relationship between ionizing radiation-induced apoptosis and stem cells in the small and large intestine. Br. J. Cancer 1998, 78, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Galli, R.; Gritti, A.; Bonfanti, L.; Vescovi, A.L. Neural stem cells: An overview. Circ. Res. 2003, 92, 598–608. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.A. Adult renal stem cells and renal repair. Curr. Opin. Nephrol. Hypertens. 2004, 13, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Yamato, E.; Taniguchi, H.; Shimoda, M.; Tashiro, F.; Hosoi, M.; Sato, T.; Fujii, S.; Miyazaki, J.I. Stimulation of cAMP signalling allows isolation of clonal pancreatic precursor cells from adult mouse pancreas. Diabetologia 2006, 49, 2359–2367. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, H.; Xu, G.; Matsumoto, S.; Kaneto, H.; Kobayashi, N.; Bonner-Weir, S.; Hayashi, S. Induction of pancreatic stem/progenitor cells into insulin-producing cells by adenoviral-mediated gene transfer technology. Cell Transplant. 2006, 15, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, H.; Kaneto, H.; Weir, G.C.; Bonner-Weir, S. PDX-1 protein containing its own antennapedia-like protein transduction domain can transduce pancreatic duct and islet cells. Diabetes 2003, 52, 1732–1737. [Google Scholar] [CrossRef] [PubMed]
- Bonner-Weir, S.; Taneja, M.; Weir, G.C.; Tatarkiewicz, K.; Song, K.H.; Sharma, A.; O’Neil, J.J. In vitro cultivation of human islets from expanded ductal tissue. Proc. Natl. Acad. Sci. USA 2000, 97, 7999–8004. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Ustinov, J.; Pulkkinen, M.A.; Lundin, K.; Korsgren, O.; Otonkoski, T. Characterization of endocrine progenitor cells and critical factors for their differentiation in human adult pancreatic cell culture. Diabetes 2003, 52, 2007–2015. [Google Scholar] [CrossRef] [PubMed]
- Heremans, Y.; Van De Casteele, M.; in’t Veld, P.; Gradwohl, G.; Serup, P.; Madsen, O.; Pipeleers, D.; Heimberg, H. Recapitulation of embryonic neuroendocrine differentiation in adult human pancreatic duct cells expressing neurogenin 3. J. Cell Biol. 2002, 159, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Nakauchi, H.; Taniguchi, H. Prospective isolation of multipotent pancreatic progenitors using flow-cytometric cell sorting. Diabetes 2004, 53, 2143–2152. [Google Scholar] [CrossRef] [PubMed]
- Oshima, Y.; Suzuki, A.; Kawashimo, K.; Ishikawa, M.; Ohkohchi, N.; Taniguchi, H. Isolation of mouse pancreatic ductal progenitor cells expressing CD133 and c-Met by flow cytometric cell sorting. Gastroenterology 2007, 132, 720–732. [Google Scholar] [CrossRef] [PubMed]
- Yatoh, S.; Dodge, R.L.; Akashi, T.; Omer, A.; Sharma, A.; Weir, G.C.; Bonner-Weir, S. Differentiation of affinity-purified human pancreatic duct cells to beta-cells. Diabetes 2007, 56, 1802–1809. [Google Scholar] [CrossRef] [PubMed]
- Seaberg, R.M.; Smukler, S.R.; Kieffer, T.J.; Enikolopov, G.; Asghar, Z.; Wheeler, M.B.; Korbutt, G.; van der Kooy, D. Clonal identification of multipotent precursors from adult mouse pancreas that generate neural and pancreatic lineages. Nat. Biotechnol. 2004, 22, 1115–1124. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, H.; Oishi, K.; Ueda, M.; Yukawa, H.; Hayashi, S.; Kobayashi, N.; Levy, M.F.; Matusmoto, S. Establishment of mouse pancreatic stem cell line. Cell Transplant. 2009, 18, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Kuise, T.; Noguchi, H.; Saitoh, I.; Kataoka, H.U.; Watanabe, M.; Noguchi, Y.; Fujiwara, T. Isolation Efficiency of Mouse Pancreatic Stem Cells Is Age Dependent. Cell Med. 2013, 5, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, H.; Naziruddin, B.; Jackson, A.; Shimoda, M.; Ikemoto, T.; Fujita, Y.; Chujo, D.; Takita, M.; Kobayashi, N.; Onaca, N.; et al. Characterization of human pancreatic progenitor cells. Cell Transplant. 2010, 19, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, H.; Bonner-Weir, S.; Wei, F.Y.; Matsushita, M.; Matsumoto, S. BETA2/NeuroD protein can be transduced into cells due to an arginine- and lysine-rich sequence. Diabetes 2005, 54, 2859–2866. [Google Scholar] [CrossRef] [PubMed]
- Nakajima-Koyama, M.; Lee, J.; Ohta, S.; Yamamoto, T.; Nishida, E. Induction of Pluripotency in Astrocytes through a Neural Stem Cell-like State. J. Biol. Chem. 2015, 290, 31173–31188. [Google Scholar] [CrossRef] [PubMed]
- Panciera, T.; Azzolin, L.; Fujimura, A.; Di Biagio, D.; Frasson, C.; Bresolin, S.; Soligo, S.; Basso, G.; Bicciato, S.; Rosato, A.; et al. Induction of Expandable Tissue-Specific Stem/Progenitor Cells through Transient Expression of YAP/TAZ. Cell Stem Cell 2016, 19, 725–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azzolin, L.; Panciera, T.; Soligo, S.; Enzo, E.; Bicciato, S.; Dupont, S.; Bresolin, S.; Frasson, C.; Basso, G.; Guzzardo, V.; et al. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell 2014, 158, 157–1570. [Google Scholar] [CrossRef] [PubMed]
- Zanconato, F.; Cordenonsi, M.; Piccolo, S. YAP/TAZ at the Roots of Cancer. Cancer Cell 2016, 29, 783–803. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, M.; Chonabayashi, K.; Nomura, M.; Tanaka, A.; Nakamura, M.; Inagaki, A.; Nishikawa, M.; Takei, I.; Oishi, A.; Tanabe, K.; et al. Epigenetic Variation between Human Induced Pluripotent Stem Cell Lines Is an Indicator of Differentiation Capacity. Cell Stem Cell 2016, 19, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Kyttälä, A.; Moraghebi, R.; Valensisi, C.; Kettunen, J.; Andrus, C.; Pasumarthy, K.K.; Nakanishi, M.; Nishimura, K.; Ohtaka, M.; Weltner, J.; et al. Genetic Variability Overrides the Impact of Parental Cell Type and Determines iPSC Differentiation Potential. Stem Cell Rep. 2016, 6, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Lee, S.; Mallard, W.; Clement, K.; Tagliazucchi, G.M.; Lim, H.; Choi, I.Y.; Ferrari, F.; Tsankov, A.M.; Pop, R.; et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat. Biotechnol. 2015, 33, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Hochedlinger, K.; Yamada, Y.; Beard, C.; Jaenisch, R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 2005, 121, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Kanagawa, O.; Nakagawa, M.; Okita, K.; Yamanaka, S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 2009, 460, 1132–1135. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, K.; Semi, K.; Yamamoto, T.; Shimizu, M.; Tanaka, A.; Mitsunaga, K.; Okita, K.; Osafune, K.; Arioka, Y.; Maeda, T.; et al. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell 2014, 156, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Miyagi-Shiohira, C.; Nakashima, Y.; Kobayashi, N.; Saitoh, I.; Watanabe, M.; Noguchi, Y.; Kinjo, T.; Noguchi, H. The development of cancer through the transient overexpression of reprogramming factors. Cell Med. in press.

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noguchi, H.; Miyagi-Shiohira, C.; Nakashima, Y. Induced Tissue-Specific Stem Cells and Epigenetic Memory in Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2018, 19, 930. https://doi.org/10.3390/ijms19040930
Noguchi H, Miyagi-Shiohira C, Nakashima Y. Induced Tissue-Specific Stem Cells and Epigenetic Memory in Induced Pluripotent Stem Cells. International Journal of Molecular Sciences. 2018; 19(4):930. https://doi.org/10.3390/ijms19040930
Chicago/Turabian StyleNoguchi, Hirofumi, Chika Miyagi-Shiohira, and Yoshiki Nakashima. 2018. "Induced Tissue-Specific Stem Cells and Epigenetic Memory in Induced Pluripotent Stem Cells" International Journal of Molecular Sciences 19, no. 4: 930. https://doi.org/10.3390/ijms19040930