ceRNA Cross-Talk in Paulownia Witches’ Broom Disease
Abstract
:1. Introduction
2. Results
2.1. Morphology of Heathy and Infected Paulownia Fortunei Seedlings
2.2. RNA Sequencing and Identification of miRNAs, mRNAs, lncRNAs, and circRNAs
2.3. Expression Analysis of miRNAs, circRNAs, lncRNAs, and mRNAs
2.4. Properties of mRNAs, lncRNAs and circRNAs
2.5. Candidate PaWB-Related miRNAs, mRNAs, lncRNAs, and circRNAs
2.6. The ceRNA Network
2.7. Verification of Seq-Results
3. Discussion
3.1. Part of the ceRNA Network Centered on pf-miR156g and pf-miR172i
3.2. Part of the ceRNA Network Centered on pf-miR8767a and pf-miR8767b
3.3. Part of the ceRNA Network Is Centered on pf-miR172h
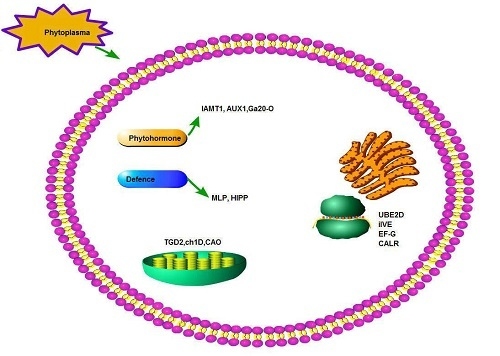
3.4. A Hypothetical Model for PaWB
4. Materials and Methods
4.1. Plant Materials
4.2. Phytoplasmas Detection
4.3. RNA Library Construction and Sequencing
4.4. Small RNA Sequencing, miRNA Identification, and the Prediction of miRNA Target Genes
4.5. Genome Mapping and Transcript Assembly
4.6. lncRNA Identification
4.7. Differential Expression Analysis of mRNAs and lncRNAs
4.8. Target Gene Prediction and Functional Analysis of lncRNAs
4.9. CircRNA Identification
4.10. Validation of ceRNAs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| lncRNA | Long noncoding RNA |
| circRNA | Circular RNA |
| miRNA | MicroRNA |
| PaWB | Paulownia witches’ broom |
| ceRNA | Competing endogenous RNA |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| CPC | Coding Potential Calculator |
| CNCI | Coding–Non-Coding Index |
| DECs | Differentially expressed circRNAs |
| DELs | Differentially expressed lncRNAs |
| DEGs | Differentially expressed genes |
| DEMs | Differentially expressed miRNA |
| AIP | Auxin-induced protein PCNT115-like |
| OPR | 12-oxophytodienoic acid reductase |
| PP2C | Probable protein phosphatase 2C 40 |
| CAD | Cinnamyl-alcohol dehydrogenase |
| ACSL | Long-chain acyl-CoA synthetase |
| Ses | Secologanin synthase-like |
| gpx | Glutathione peroxidase |
| CAO | Chlorophyllide a oxygenase |
| Ga20-O | Gibberellin 20-oxidase |
| AUX1 | Auxin influx carrier |
| UBE2D | Ubiquitin-conjugating enzyme E2 D |
| ilvE | Branchedchain amino acid aminotransferase |
| GsSRK | G-type lectin S-receptor-like serine/threonine-protein kinase |
| EF-G | Elongation factor G |
| CALR | Calreticulin |
| CASPL | CASP-like protein 2B1 |
| TGD2 | Protein TRIGALACTOSYLDIACYLGLYCEROL 2 |
| GA | Gibberellin |
| ch1D | Magnesium chelatase subunit D |
| PHO | Acid phosphatase |
| MLPs | Major latex proteins |
| IAA | Indole-3-acetic acid |
| IAMT1 | IAA-methyltransferase-1 |
| HIPPs | Heavy metal-associated isoprenylated plant proteins |
References
- Jiang, Z.F.; Huang, S.Z.; Han, Y.L.; Zhao, J.Z.; Fu, J.J. Physiological response of Cu and Cu mine tailing remediation of Paulownia fortunei (seem) hemsl. Ecotoxicology 2012, 21, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Fan, G.; Dong, Y.; Zhai, X.; Deng, M.; Zhao, Z.; Liu, W.; Cao, Y. Implications of polyploidy events on the phenotype, microstructure, and proteome of Paulownia australis. PLoS ONE 2017, 12, e0172633. [Google Scholar] [CrossRef] [PubMed]
- Yadav, N.K.; Vaidya, B.N.; Henderson, K.; Lee, J.F.; Stewart, W.M.; Dhekney, S.A.; Joshee, N. A review of Paulownia biotechnology: A short rotation, fast growing multipurpose bioenergy tree. Am. J. Plant Sci. 2013, 4, 2070–2082. [Google Scholar] [CrossRef]
- Lee, I.M.; Davis, R.E.; Gundersen-Rindal, D.E. Phytoplasma: Phytopathogenic mollicutes. Annu. Rev. Microbiol. 2000, 54, 221–255. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Dong, Y.; Deng, M.; Zhao, Z.; Niu, S.; Xu, E. Plant–pathogen interaction, circadian rhythm, and hormone-related gene expression provide indicators of phytoplasma infection in Paulownia fortunei. Int. J. Mol. Sci. 2014, 15, 23141–23162. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Xu, E.; Deng, M.; Zhao, Z.; Niu, S. Phenylpropanoid metabolism, hormone biosynthesis and signal transduction-related genes play crucial roles in the resistance of Paulownia fortunei to paulownia witches’ broom phytoplasma infection. Genes Genom. 2015, 37, 913–929. [Google Scholar] [CrossRef]
- Liu, R.; Dong, Y.; Fan, G.; Zhao, Z.; Deng, M.; Cao, X.; Niu, S. Discovery of genes related to witches broom disease in Paulownia tomentosa × Paulownia fortunei by a de novo assembled transcriptome. PLoS ONE 2013, 8, e80238. [Google Scholar] [CrossRef] [PubMed]
- Mou, H.Q.; Lu, J.; Zhu, S.F.; Lin, C.L.; Tian, G.Z.; Xu, X.; Zhao, W.J. Transcriptomic analysis of Paulownia infected by paulownia witches’-broom phytoplasma. PLoS ONE 2013, 8, e77217. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Cao, X.; Zhao, Z.; Deng, M. Transcriptome analysis of the genes related to the morphological changes of Paulownia tomentosa plantlets infected with phytoplasma. Acta Physiol. Plant. 2015, 37, 1–12. [Google Scholar] [CrossRef]
- Fan, G.; Cao, X.; Niu, S.; Deng, M.; Zhao, Z.; Dong, Y. Transcriptome, microRNA, and degradome analyses of the gene expression of paulownia with phytoplamsa. BMC Genom. 2015, 16, 896. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Niu, S.; Zhao, Z.; Deng, M.; Xu, E.; Wang, Y.; Yang, L. Identification of microRNAs and their targets in Paulownia fortunei plants free from phytoplasma pathogen after methyl methane sulfonate treatment. Biochimie 2016, 127, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Fan, G.; Deng, M.; Zhao, Z.; Xu, E.; Cao, L. Discovery of microRNAs and transcript targets related to witches’ broom disease in Paulownia fortunei by high-throughput sequencing and degradome approach. Mol. Genet. Genom. 2016, 291, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Cao, Y.; Deng, M.; Zhai, X.; Zhao, Z.; Niu, S.; Ren, Y. Identification and dynamic expression profiling of microRNAs and target genes of Paulownia tomentosa in response to paulownia witches’ broom disease. Acta Physiol. Plant. 2016, 39, 28. [Google Scholar] [CrossRef]
- Fan, G.; Niu, S.; Xu, T.; Deng, M.; Zhao, Z.; Wang, Y.; Cao, L.; Wang, Z. Plant–pathogen interaction-related micrornas and their targets provide indicators of phytoplasma infection in Paulownia tomentosa × Paulownia fortunei. PLoS ONE 2015, 10, e0140590. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, W.; Fan, G.; Zhai, X.; Zhao, Z.; Dong, Y.; Deng, M.; Cao, Y. Quantitative proteome-level analysis of paulownia witches’ broom disease with methyl methane sulfonate assistance reveals diverse metabolic changes during the infection and recovery processes. PeerJ 2017, 5, e3495. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Wang, Z.; Li, X.; Zhao, Z.; Deng, M.; Dong, Y.; Cao, X.; Fan, G. Comparative proteomic analysis of Paulownia fortunei response to phytoplasma infection with dimethyl sulfate treatment. Int. J. Genom. 2017, 2017, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Fan, G.; Dong, Y.; Zhao, Z.; Deng, M.; Wang, Z.; Liu, W. Proteome profiling of paulownia seedlings infected with phytoplasma. Front. Plant Sci. 2017, 8, 342. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zhai, X.; Deng, M.; Zhao, Z.; Fan, G. Relationship between metabolites variation and paulownia witches’broom. Sci. Silvae Sin. 2017, 53, 85–93. [Google Scholar]
- Wang, Z.; Zhai, X.; Cao, Y.; Dong, Y.; Fan, G. Long non-coding RNAs responsive to witches’ broom disease in Paulownia tomentosa. Forests 2017, 8, 348. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Hao, Z.; Yan, J.; Li, G. Genome-wide identification and functional analysis of lincRNAs acting as miRNA targets or decoys in maize. BMC Genom. 2015, 16, 793. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.W.; Zhou, X.H.; Wang, R.R.; Peng, W.L.; An, Y.; Chen, L.L. Functional analysis of long intergenic non-coding RNAs in phosphate-starved rice using competing endogenous RNA network. Sci. Rep. 2016, 6, 20715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Wang, C.; Bao, H.; Chen, H.; Wang, Y. Genome-wide identification and characterization of novel lncRNAs in populus under nitrogen deficiency. Mol. Genet. Genom. 2016, 291, 1663–1680. [Google Scholar] [CrossRef] [PubMed]
- Gai, Y.P.; Li, Y.Q.; Guo, F.Y.; Yuan, C.Z.; Mo, Y.Y.; Zhang, H.L.; Wang, H.; Ji, X.L. Analysis of phytoplasma-responsive sRNAs provide insight into the pathogenic mechanisms of mulberry yellow dwarf disease. Sci. Rep. 2014, 4, 5378. [Google Scholar] [CrossRef] [PubMed]
- Ehya, F.; Monavarfeshani, A.; Mohseni Fard, E.; Karimi Farsad, L.; Khayam Nekouei, M.; Mardi, M.; Salekdeh, G.H. Phytoplasma-responsive microRNAs modulate hormonal, nutritional, and stress signalling pathways in Mexican lime trees. PLoS ONE 2013, 8, e66372. [Google Scholar] [CrossRef] [PubMed]
- Shao, F.; Zhang, Q.; Liu, H.; Lu, S.; Qiu, D. Genome-wide identification and analysis of microRNAs involved in witches’-broom phytoplasma response in ziziphus jujuba. PLoS ONE 2016, 11, e0166099. [Google Scholar] [CrossRef] [PubMed]
- Niu, S. Changes of Omics after Paulownia Trees Infected by the Paulownia Witches Phytoplasma Pathogen. Ph.D. Thesis, Henan Agricultural University, Zhengzhou, China, 2017. [Google Scholar]
- Liao, W.; Chen, Z.; Ye, M.; Ma, H.; Gao, K.; Lei, B.; An, X. Prediction and bioinformatics analysis of miRNA target genes in Populus tomentosa under fungus stress. Chin. J. Cell Biol. 2014, 36, 1506–1513. [Google Scholar]
- Mardi, M.; Karimi Farsad, L.; Gharechahi, J.; Salekdeh, G.H. In-depth transcriptome sequencing of Mexican lime trees infected with Candidatus phytoplasma aurantifolia. PLoS ONE 2015, 10, e0130425. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Bai, T.; Li, Y.; Ruan, X.; Li, H. Proteomic analysis of fusarium oxysporum f. sp. Cubense tropical race 4-inoculated response to fusarium wilts in the banana root cells. Proteome Sci. 2013, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- Margaria, P.; Abba, S.; Palmano, S. Novel aspects of grapevine response to phytoplasma infection investigated by a proteomic and phospho-proteomic approach with data integration into functional networks. BMC Genom. 2013, 14, 38. [Google Scholar] [CrossRef] [PubMed]
- Stintzi, A.; Browse, J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 10625–10630. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, G.K.; Jwa, N.S.; Shibato, J.; Han, O.; Iwahashi, H.; Rakwal, R. Diverse environmental cues transiently regulate OsOPR1 of the “octadecanoid pathway” revealing its importance in rice defense/stress and development. Biochem. Biophys. Res. Commun. 2003, 310, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Schweighofer, A.; Hirt, H.; Meskiene, I. Plant pp2c phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 2004, 9, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Irmler, S.; Schröder, G.; St-Pierre, B.; Crouch, N.P.; Hotze, M.; Schmidt, J.; Strack, D.; Matern, U.; Schröder, J. Indole alkaloid biosynthesis in Catharanthus roseus: New enzyme activities and identification of cytochrome P450 CYP72A1 as secologanin synthase. Plant J. 2000, 24, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.R. The glutathione peroxidases. Cell. Mol. Life Sci. 2001, 57, 1825–1835. [Google Scholar] [CrossRef]
- Oster, U.; Tanaka, R.; Tanaka, A.; Rudiger, W. Cloning and functional expression of the gene encoding the key enzyme for chlorophyll b biosynthesis (CAO) from Arabidopsis thaliana. Plant J. 2000, 21, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, M.; Wei, S.; Qin, F.; Zhao, H.; Suo, B. Identification of circular RNAs and their targets in leaves of Triticum aestivum L. Under dehydration stress. Front. Plant Sci. 2016, 7, 2024. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yu, F.; Wu, W.; Zhang, Y.; Chang, W.; Ponnusamy, M.; Wang, K.; Li, P. Circular RNAs: A novel type of non-coding RNA and their potential implications in antiviral immunity. Int. J. Biol. Sci. 2017, 13, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Phinney, B.O. Gibberellin A_1, dwarfism and the control of shoot elongation in higher plants. In The Biosynthesis and Metabolism of Plant Hormones. Society for Experimental Biology Seminar Series; Cambridge University Press: Cambridge, UK, 1984; Volume 23, pp. 17–41. [Google Scholar]
- Evans, M.L.; Cleland, R.E. The action of auxin on plant cell elongation. CRC Rev. Plant Sci. 1985, 2, 317–365. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.H.; Li, D.M.; Yin, M.H.; Li, X.B.; Zhang, M.; Wang, Y.J.; Dong, J.; Zhao, J.; Luo, M.; Luo, X.Y.; et al. Gibberellin 20-oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. J. Plant Physiol. 2010, 167, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Van Minnebruggen, A.; Neyt, P.; de Groeve, S.; Coussens, G.; Ponce, M.R.; Micol, J.L.; van Lijsebettens, M. The ang3 mutation identified the ribosomal protein gene rpl5b with a role in cell expansion during organ growth. Physiol. Plant 2010, 138, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.G.; Selmer, M.; Dunham, C.M.; Weixlbaumer, A.; Kelley, A.C.; Ramakrishnan, V. The structure of the ribosome with elongation factor g trapped in the posttranslocational state. Science 2009, 326, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Michalak, M.; Groenendyk, J.; Szabo, E.; Gold, L.I.; Opas, M. Calreticulin, a multi-process calcium-buffering chaperone of the endoplasmic reticulum. Biochem. J. 2009, 417, 651–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, X.; Gai, Y.; Zheng, C.; Mu, Z. Comparative proteomic analysis provides new insights into mulberry dwarf responses in mulberry (Morus alba L.). Proteomics 2009, 9, 5328–5339. [Google Scholar] [CrossRef] [PubMed]
- Monavarfeshani, A.; Mirzaei, M.; Sarhadi, E.; Amirkhani, A.; Khayam Nekouei, M.; Haynes, P.A.; Mardi, M.; Salekdeh, G.H. Shotgun proteomic analysis of the Mexican lime tree infected with “candidatusPhytoplasma aurantifolia”. J. Proteome Res. 2013, 12, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Awai, K.; Xu, C.; Tamot, B.; Benning, C. A phosphatidic acid-binding protein of the chloroplast inner envelope membrane involved in lipid trafficking. Proc. Natl. Acad. Sci. USA 2006, 103, 10817–10822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osmark, P.; Boyle, B.; Brisson, N. Sequential and structural homology between intracellular pathogenesis-related proteins and a group of latex proteins. Plant Mol. Biol. 1998, 38, 1243–1246. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hou, X.; Tsuge, T.; Ding, M.; Aoyama, T.; Oka, A.; Gu, H.; Zhao, Y.; Qu, L.J. The possible action mechanisms of indole-3-acetic acid methyl ester in Arabidopsis. Plant Cell Rep. 2008, 27, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Ferrer, J.L.; Ross, J.; Guan, J.; Yang, Y.; Pichersky, E.; Noel, J.P.; Chen, F. Structural, biochemical, and phylogenetic analyses suggest that indole-3-acetic acid methyltransferase is an evolutionarily ancient member of the sabath family. Plant Physiol. 2008, 146, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Gai, Y.P.; Han, X.J.; Li, Y.Q.; Yuan, C.Z.; Mo, Y.Y.; Guo, F.Y.; Liu, Q.X.; Ji, X.L. Metabolomic analysis reveals the potential metabolites and pathogenesis involved in mulberry yellow dwarf disease. Plant Cell Environ. 2014, 37, 1474–1490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Abreu-Neto, J.B.; Turchetto-Zolet, A.C.; de Oliveira, L.F.; Zanettini, M.H.; Margis-Pinheiro, M. Heavy metal-associated isoprenylated plant protein (HIPP): Characterization of a family of proteins exclusive to plants. FEBS J. 2013, 280, 1604–1616. [Google Scholar] [CrossRef] [PubMed]
- Tai, C.F.; Lin, C.P.; Sung, Y.C.; Chen, J.C. Auxin influences symptom expression and phytoplasma colonisation in periwinkle infected with periwinkle leaf yellowing phytoplasma. Ann. Appl. Biol. 2013, 163, 420–429. [Google Scholar] [CrossRef]
- Lee, I.M.; Hammond, R.W.; Davis, R.E.; Gundersen, D.E. Universal amplification and analysis of pathogen 16s rDNA for classification and identification of mycoplasmalike organisms. Phytopathology 1993, 83, 834–842. [Google Scholar] [CrossRef]
- Fan, G.; Zhang, S.; Zhai, X.; Liu, F.; Dong, Z. Effects of antibiotics on the paulownia witches’ broom phytoplasmas and pathogenic protein related to witches’ broom symptom. Sci. Silvae Sin. 2007, 43, 138–142. [Google Scholar]
- Sun, W.; Xu, X.H.; Wu, X.; Wang, Y.; Lu, X.; Sun, H.; Xie, X. Genome-wide identification of microRNAs and their targets in wild type and phyB mutant provides a key link between microRNAs and the phyB-mediated light signaling pathway in rice. Front. Plant Sci. 2015, 6, 372. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012, 9, 357. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. Tophat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. Stringtie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Frazee, A.C.; Pertea, G.; Jaffe, A.E.; Langmead, B.; Salzberg, S.L.; Leek, J.T. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 2015, 33, 243–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. Cpc: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Salzberg, S.L. Tophat-fusion: An algorithm for discovery of novel fusion transcripts. Genome Biol. 2011, 12, R72. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Dong, R.; Zhang, Y.; Zhang, J.L.; Luo, Z.; Zhang, J.; Chen, L.L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef] [PubMed]
- Hang, R.; Liu, C.; Ahmad, A.; Zhang, Y.; Lu, F.; Cao, X. Arabidopsis protein arginine methyltransferase 3 is required for ribosome biogenesis by affecting precursor ribosomal RNA processing. Proc. Natl. Acad. Sci. USA 2014, 111, 16190–16195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hang, R.; Deng, X.; Liu, C.; Mo, B.; Cao, X. Circular RT-PCR assay using Arabidopsis samples. Bio-Protocol 2015, 5, e1533. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, G.; Wang, Z.; Zhai, X.; Cao, Y. ceRNA Cross-Talk in Paulownia Witches’ Broom Disease. Int. J. Mol. Sci. 2018, 19, 2463. https://doi.org/10.3390/ijms19082463
Fan G, Wang Z, Zhai X, Cao Y. ceRNA Cross-Talk in Paulownia Witches’ Broom Disease. International Journal of Molecular Sciences. 2018; 19(8):2463. https://doi.org/10.3390/ijms19082463
Chicago/Turabian StyleFan, Guoqiang, Zhe Wang, Xiaoqiao Zhai, and Yabing Cao. 2018. "ceRNA Cross-Talk in Paulownia Witches’ Broom Disease" International Journal of Molecular Sciences 19, no. 8: 2463. https://doi.org/10.3390/ijms19082463