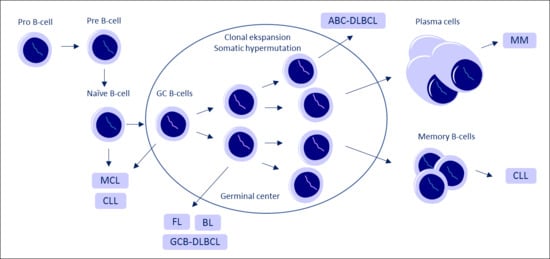
Long Non-Coding RNAs Guide the Fine-Tuning of Gene Regulation in B-Cell Development and Malignancy
Abstract
:1. Introduction
2. LncRNA in B-Cell Development
3. Long Non-Coding RNA Expression in Various B-Cell Malignancies
3.1. Acute Lymphoblastic Leukemia
3.2. Chronic Lymphocytic Leukemia
3.3. Mantle Cell Lymphoma
3.4. Diffuse Large B-Cell Lymphoma and Follicular Lymphoma
3.5. Burkitt’s Lymphoma
3.6. Multiple Myeloma
4. Circular RNA in B-Cell Development and Malignancies
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABC-DLBCL | Activated B-cell diffuse large B-cell lymphoma |
| ALL | Acute lymphoblastic leukemia |
| ANRIL | Antisense non-coding RNA in the INK4-ARF locus |
| B-ALL | B-Cell Acute lymphoblastic leukemia |
| BALR | B-ALL-associated long non-coding RNAs |
| BL | Burkitt Lymphoma |
| CCAT1 | Colon cancer associated transcript 1 |
| ceRNA | Competing endogenous RNA |
| cGAS | Cyclic GMP-AMP synthase |
| CRISPR | Clustered regularly interspaced palindromic repeats |
| CRNDE | Colorectal neoplasia differentially expressed |
| circRNA | Circular RNA |
| ciRS-7 | Circular RNA sponge for miR-7 |
| CLL | Chronic lymphocytic leukemia |
| DLBCL | Diffuse large B-cell lymphoma |
| DzNep | 3-deazanoplanocin A |
| FFPE | Formalin-fixed paraffin-embedded |
| FL | Follicular lymphoma |
| GAS5 | Growth specific 5 |
| GC | Germinal center |
| GCB-DLBCL | Germinal center diffuse large B-cell lymphoma |
| GEO | Gene expression omnibus |
| HOTAIR | The HOX transcript antisense intergenic RNA |
| IPI | International prognostic index |
| KD | Knockdown |
| lincRNA-p21 | Large intergenic non-coding RNA p21 |
| lncRNA | Long non-coding RNA |
| LT-HSCs | Long term hematopoietic stem cells |
| MALAT1 | Metastasis associated lung adenocarcinoma transcript 1 |
| MCL | Mantle cell lymphoma |
| MIAT | Myocardial infarction associated transcript |
| MINCR | MYC-induced long non-coding RNA |
| miRNA | MicroRNA |
| MEG3 | Maternally expressed 3 |
| MGUS | Monoclonal gammopathy of undetermined significance |
| MM | Maternally expressed 3 |
| NEAT1 | Nuclear enriched abundant transcript 1 |
| OS | Overall survival |
| PAIR | PAX5 activated intergenic repeat |
| PANDA | P21 associated ncRNA DNA damage activated |
| PCAT1 | Prostate cancer associated transcript 1 |
| PEG10 | Paternally expressed 10 |
| PFS | Progression-free survival |
| PCL | Plasma cell leukemia |
| PDIA3P | Protein disulfide isomerase family A member 3 pseudogene 1 |
| PRC2 | Polycomb repressive complex 2 |
| RNAi | RNA interference |
| RNA-seq | RNA sequencing |
| rRNA | Ribosomal RNA |
| SMM | Smoldering multiple myeloma |
| STAiRs | STAT3-induced lncRNAs |
| treRNA1 | Translation regulatory long non-coding RNA1 |
| TUG1 | Taurine upregulated 1 |
| UCA1 | Urothelial cancer associated 1 |
References
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigó, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S.; et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, M.W. A short guide to long non-coding RNA gene nomenclature. Hum. Genom. 2014, 8, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- St. Laurent, G.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Cawley, S.; Bekiranov, S.; Ng, H.H.; Kapranov, P.; Sekinger, E.A.; Kampa, D.; Piccolboni, A.; Sementchenko, V.; Cheng, J.; Williams, A.J.; et al. Unbiased Mapping of Transcription Factor Binding Sites along Human Chromosomes 21 and 22 Points to Widespread Regulation of Noncoding RNAs. Cell 2004, 116, 499–509. [Google Scholar] [CrossRef]
- Ponjavic, J.; Ponting, C.P.; Lunter, G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007, 17, 556–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimitrova, N.; Zamudio, J.R.; Jong, R.M.; Soukup, D.; Resnick, R.; Sarma, K.; Ward, A.J.; Raj, A.; Lee, J.T.; Sharp, P.A.; et al. LincRNA-p21 Activates p21 In cis to Promote Polycomb Target Gene Expression and to Enforce the G1/S Checkpoint. Mol. Cell 2014, 54, 777–790. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An Architectural Role for a Nuclear Non-coding RNA: NEAT1 RNA is Essential for the Structure of Paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Ebralidze, A.K.; Guibal, F.C.; Steidl, U.; Zhang, P.; Lee, S.; Bartholdy, B.; Jorda, M.A.; Petkova, V.; Rosenbauer, F.; Huang, G.; et al. PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev. 2008, 22, 2085–2092. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Lou, Z.; Gupta, M. The Long Non-Coding RNA GAS5 Cooperates with the Eukaryotic Translation Initiation Factor 4E to Regulate c-myc Translation. PLoS ONE 2014, 9, e107016. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. CircRNA Biogenesis competes with Pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelmohsen, K.; Panda, A.C.; Munk, R.; Grammatikakis, I.; Dudekula, D.B.; De, S.; Kim, J.; Noh, J.H.; Kim, K.M.; Martindale, J.L.; et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017, 14, 361–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Gao, X.; Zhang, M.; Yan, S.; Sun, C.; Xiao, F.; Huang, N.; Yang, X.; Zhao, K.; Zhou, H.; et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J. Natl. Cancer Inst. 2018, 110, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Stagsted, L.V.W.; Nielsen, K.M.; Daugaard, I.; Hansen, T.B. Non-coding AUG circRNAs constitute an abundant and conserved subclass of circles. bioRxiv, 2018. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Okholm, T.L.H.; Venø, M.T.; Kjems, J. Circular RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol. 2017, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, J.; Wei, X.; Song, C.; Dong, D.; Huang, Y.; Lan, X.; Plath, M.; Lei, C.; Ma, Y.; et al. CircFUT10 reduces proliferation and facilitates differentiation of myoblasts by sponging miR-133a. J. Cell. Physiol. 2018, 223, 4643–4651. [Google Scholar] [CrossRef] [PubMed]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Venø, M.T.; Hansen, T.B.; Venø, S.T.; Clausen, B.H.; Grebing, M.; Finsen, B.; Holm, I.E.; Kjems, J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015, 16, 245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greene, J.; Baird, A.-M.; Brady, L.; Lim, M.; Gray, S.G.; McDermott, R.; Finn, S.P. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front. Mol. Biosci. 2017, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Hansen, T.B.; Venø, M.T.; Kjems, J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 2017, 37, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Diederichs, S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012, 9, 703–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Küppers, R.; Klein, U.; Hansmann, M.L.; Rajewsky, K. Cellular origin of human B-cell lymphomas. N. Engl. J. Med. 1999, 341, 1520–1529. [Google Scholar] [CrossRef] [PubMed]
- Bonnal, R.J.P.; Ranzani, V.; Arrigoni, A.; Curti, S.; Panzeri, I.; Gruarin, P.; Abrignani, S.; Rossetti, G.; Pagani, M. De novo transcriptome profiling of highly purified human lymphocytes primary cells. Sci. Data 2015, 2, 150051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casero, D.; Sandoval, S.; Seet, C.S.; Scholes, J.; Zhu, Y.; Ha, V.L.; Luong, A.; Parekh, C.; Crooks, G.M. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat. Immunol. 2015, 16, 1282–1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petri, A.; Dybkær, K.; Bøgsted, M.; Thrue, C.A.; Hagedorn, P.H.; Schmitz, A.; Bødker, J.S.; Johnsen, H.E.; Kauppinen, S. Long Noncoding RNA Expression during Human B-Cell Development. PLoS ONE 2015, 10, e0138236. [Google Scholar] [CrossRef] [PubMed]
- Ranzani, V.; Rossetti, G.; Panzeri, I.; Arrigoni, A.; Bonnal, R.J.P.; Curti, S.; Gruarin, P.; Provasi, E.; Sugliano, E.; Marconi, M.; et al. LincRNA landscape in human lymphocytes highlights regulation of T cell differentiation by linc-MAF-4. Nat. Immunol. 2015, 16, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Tayari, M.M.; Winkle, M.; Kortman, G.; Sietzema, J.; de Jong, D.; Terpstra, M.; Mestdagh, P.; Kroese, F.G.M.; Visser, L.; Diepstra, A.; et al. Long Noncoding RNA Expression Profiling in Normal B-Cell Subsets and Hodgkin Lymphoma Reveals Hodgkin and Reed-Sternberg Cell–Specific Long Noncoding RNAs. Am. J. Pathol. 2016, 186, 2462–2472. [Google Scholar] [CrossRef] [PubMed]
- Brazao, T.F.; Johnson, J.S.; Muller, J.; Heger, A.; Ponting, C.P.; Tybulewicz, V.L. Long noncoding RNAs in B-cell development and activation. Blood 2016, 128, e10–e19. [Google Scholar] [CrossRef] [PubMed]
- Nutt, S.L.; Heavey, B.; Rolink, A.G.; Busslinger, M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 1999, 401, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Verma-Gaur, J.; Torkamani, A.; Schaffer, L.; Head, S.R.; Schork, N.J.; Feeney, A.J. Noncoding transcription within the Igh distal VH region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc. Natl. Acad. Sci. USA 2012, 109, 17004–17009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Schmidt-Supprian, M.; Shi, Y.; Hobeika, E.; Barteneva, N.; Jumaa, H.; Pelanda, R.; Reth, M.; Skok, J.; Rajewsky, K. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev 2007, 21, 1179–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syrett, C.M.; Sindhava, V.; Hodawadekar, S.; Myles, A.; Liang, G.; Zhang, Y.; Nandi, S.; Cancro, M.; Atchison, M.; Anguera, M.C. Loss of Xist RNA from the inactive X during B cell development is restored in a dynamic YY1-dependent two-step process in activated B cells. PLoS Genet. 2017, 13, e1007050. [Google Scholar] [CrossRef] [PubMed]
- DeKoter, R.P.; Singh, H. Regulation of B Lymphocyte and Macrophage Development by Graded Expression of PU.1. Science 2000, 288, 1439–1441. [Google Scholar] [CrossRef] [PubMed]
- Rosenbauer, F.; Owens, B.M.; Yu, L.; Tumang, J.R.; Steidl, U.; Kutok, J.L.; Clayton, L.K.; Wagner, K.; Scheller, M.; Iwasaki, H.; et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat. Genet. 2006, 38, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Fang, K.; Han, B.-W.; Chen, Z.-H.; Lin, K.-Y.; Zeng, C.-W.; Li, X.-J.; Li, J.-H.; Luo, X.-Q.; Chen, Y.-Q. A distinct set of long non-coding RNAs in childhood MLL-rearranged acute lymphoblastic leukemia: Biology and epigenetic target. Hum. Mol. Genet. 2014, 23, 3278–3288. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.H.; Behm, F.G.; Downing, J.R.; Hancock, M.L.; Shurtleff, S.A.; Ribeiro, R.C.; Head, D.R.; Mahmoud, H.H.; Sandlund, J.T.; Furman, W.L. 11q23/MLL rearrangement confers a poor prognosis in infants with acute lymphoblastic leukemia. J. Clin. Oncol. 1994, 12, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Fernando, T.R.; Rodriguez-Malave, N.I.; Waters, E.V.; Yan, W.; Casero, D.; Basso, G.; Pigazzi, M.; Rao, D.S. LncRNA Expression Discriminates Karyotype and Predicts Survival in B-Lymphoblastic Leukemia. Mol. Cancer Res. 2015, 13, 839–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Malavé, N.I.; Fernando, T.R.; Patel, P.C.; Contreras, J.R.; Palanichamy, J.K.; Tran, T.M.; Anguiano, J.; Davoren, M.J.; Alberti, M.O.; Pioli, K.T.; et al. BALR-6 regulates cell growth and cell survival in B-lymphoblastic leukemia. Mol. Cancer 2015, 14, 214. [Google Scholar] [CrossRef]
- Fernando, T.R.; Contreras, J.R.; Zampini, M.; Rodriguez-Malave, N.I.; Alberti, M.O.; Anguiano, J.; Tran, T.M.; Palanichamy, J.K.; Gajeton, J.; Ung, N.M.; et al. The lncRNA CASC15 regulates SOX4 expression in RUNX1-rearranged acute leukemia. Mol. Cancer 2017. [Google Scholar] [CrossRef] [PubMed]
- Ghazavi, F.; De Moerloose, B.; Van Loocke, W.; Wallaert, A.; Helsmoortel, H.H.; Ferster, A.; Bakkus, M.; Plat, G.; Delabesse, E.; Uyttebroeck, A.; et al. Unique long non-coding RNA expression signature in ETV6/RUNX1-driven B-cell precursor acute lymphoblastic leukemia. Oncotarget 2016, 7, 73769–73780. [Google Scholar] [CrossRef] [PubMed]
- Moorman, A.V.; Ensor, H.M.; Richards, S.M.; Chilton, L.; Schwab, C.; Kinsey, S.E.; Vora, A.; Mitchell, C.D.; Harrison, C.J. Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: Results from the UK Medical Research Council ALL97/99 randomised trial. Lancet Oncol. 2010, 11, 429–438. [Google Scholar] [CrossRef]
- Lajoie, M.; Drouin, S.; Caron, M.; St-Onge, P.; Ouimet, M.; Gioia, R.; Lafond, M.-H.; Vidal, R.; Richer, C.; Oualkacha, K.; et al. Specific expression of novel long non-coding RNAs in high-hyperdiploid childhood acute lymphoblastic leukemia. PLoS ONE 2017, 12, e0174124. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sehgal, L.; Jain, N.; Khashab, T.; Mathur, R.; Samaniego, F. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J. Transl. Med. 2016, 14, 346. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-J.; Chai, Y.; Guo, X.-J.; Chu, S.-L.; Zhang, L.-S. The effects of the long non-coding RNA MALAT-1 regulated autophagy-related signaling pathway on chemotherapy resistance in diffuse large B-cell lymphoma. Biomed. Pharmacother. 2017, 89, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Ronchetti, D.; Agnelli, L.; Taiana, E.; Galletti, S.; Manzoni, M.; Todoerti, K.; Musto, P.; Strozzi, F.; Neri, A. Distinct lncRNA transcriptional fingerprints characterize progressive stages of multiple myeloma. Oncotarget 2016, 7, 14814–14830. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.-F.; Chang, Y.C.; Chang, C.-S.; Lin, S.-F.; Liu, Y.-C.; Hsiao, H.-H.; Chang, J.-G.; Liu, T.-C. MALAT1 long non-coding RNA is overexpressed in multiple myeloma and may serve as a marker to predict disease progression. BMC Cancer 2014, 14, 809. [Google Scholar] [CrossRef] [PubMed]
- Blume, C.J.; Hotz-Wagenblatt, A.; Hüllein, J.; Sellner, L.; Jethwa, A.; Stolz, T.; Slabicki, M.; Lee, K.; Sharathchandra, A.; Benner, A.; et al. p53-dependent non-coding RNA networks in chronic lymphocytic leukemia. Leukemia 2015, 29, 2015–2023. [Google Scholar] [CrossRef] [PubMed]
- Isin, M.; Ozgur, E.; Cetin, G.; Erten, N.; Aktan, M.; Gezer, U.; Dalay, N. Investigation of circulating lncRNAs in B-cell neoplasms. Clin. Chim. Acta 2014, 431, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Yilan, W.; Han, W. LncRNA NEAT1 promotes dexamethasone resistance in multiple myeloma by targeting miR-193a/MCL1 pathway. J. Biochem. Mol. Toxicol. 2017, 32, e22008. [Google Scholar] [CrossRef]
- Sattari, A.; Siddiqui, H.; Moshiri, F.; Ngankeu, A.; Nakamura, T.; Kipps, T.J.; Croce, C.M. Upregulation of long noncoding RNA MIAT in aggressive form of chronic lymphocytic leukemias. Oncotarget 2016, 7, 54174–54182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sone, M.; Hayashi, T.; Tarui, H.; Agata, K.; Takeichi, M.; Nakagawa, S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 2007, 120, 2498–2506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronchetti, D.; Agnelli, L.; Pietrelli, A.; Todoerti, K.; Manzoni, M.; Taiana, E.; Neri, A. A compendium of long non-coding RNAs transcriptional fingerprint in multiple myeloma. Sci. Rep. 2018, 8, 6557. [Google Scholar] [CrossRef] [PubMed]
- Subhash, S.; Andersson, P.-O.; Kosalai, S.T.; Kanduri, C.; Kanduri, M. Global DNA methylation profiling reveals new insights into epigenetically deregulated protein coding and long noncoding RNAs in CLL. Clin. Epigenet. 2016, 8, 106. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, Y.-B.; He, X.; Huang, Y.-F.; Wu, Q.-N.; Zhou, Y.-C.; Hao, D.-J. Long Noncoding RNA CRNDE Promotes Multiple Myeloma Cell Growth by Suppressing miR-451. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.-Y.; Wu, B.; Yan, W.; Gong, Z.-M.; Sun, Q.; Wang, H.-H.; Yang, W. Microarray expression profiles of long non-coding RNAs in germinal center-like diffuse large B-cell lymphoma. Oncol. Rep. 2017, 38, 1363–1372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, D.; Fang, C.; Li, X.; Geng, Y.; Li, R.; Wu, C.; Jiang, J.; Wu, C. Predictive analysis of long non-coding RNA expression profiles in diffuse large B-cell lymphoma. Oncotarget 2017, 8, 23228–23236. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Han, J.; Li, Z.; Yang, H.; Sui, Y.; Wang, M. Elevated RNA expression of long non-coding HOTAIR promotes cell proliferation and predicts a poor prognosis in patients with diffuse large B cell lymphoma. Mol. Med. Rep. 2016, 13, 5125–5131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, E.J.; Kim, S.H.; Yang, W.I.; Ko, Y.H.; Yoon, S.O. Long Non-coding RNA HOTAIR Expression in Diffuse Large B-Cell Lymphoma: In Relation to Polycomb Repressive Complex Pathway Proteins and H3K27 Trimethylation. J. Pathol. Transl. Med. 2016, 50, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mourtada-Maarabouni, M.; Williams, G.T. Role of GAS5 noncoding RNA in mediating the effects of rapamycin and its analogues on mantle cell lymphoma cells. Clin. Lymphoma Myeloma Leuk. 2014, 14, 468–473. [Google Scholar] [CrossRef] [PubMed]
- Lenka, S.; Barbora, G.; Veronika, K.; Lenka, R.; Jana, F.; Jiri, J.; Lucie, B.; Roberta, V.; Martina, A.; Miroslav, P.; et al. Deregulated expression of long non-coding RNA UCA1 in multiple myeloma. Eur. J. Haematol. 2017, 99, 223–233. [Google Scholar] [CrossRef]
- Zhang, Z.-S.; Wang, J.; Zhu, B.-Q.; Ge, L. Long noncoding RNA UCA1 promotes multiple myeloma cell growth by targeting TGF-β. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 1374–1379. [Google Scholar] [PubMed]
- Pan, Y.; Chen, H.; Shen, X.; Wang, X.; Ju, S.; Lu, M.; Cong, H. Serum level of long noncoding RNA H19 as a diagnostic biomarker of multiple myeloma. Clin. Chim. Acta 2018, 480, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Pan, J.; Zhang, N.; Wei, W.; Yu, S.; Ai, L. Knockdown of long non-coding RNA H19 inhibits multiple myeloma cell growth via NF-κB pathway. Sci. Rep. 2017, 7, 18079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouimet, M.; Drouin, S.; Lajoie, M.; Caron, M.; St-Onge, P.; Gioia, R.; Richer, C.; Sinnett, D. A childhood acute lymphoblastic leukemia-specific lncRNA implicated in prednisolone resistance, cell proliferation, and migration. Oncotarget 2017, 8, 7477–7488. [Google Scholar] [CrossRef] [PubMed]
- Arthur, G.; Almamun, M.; Taylor, K. Hypermethylation of antisense long noncoding RNAs in acute lymphoblastic leukemia. Epigenomics 2017, 9, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, Y.; Segura, V.; Marín-Béjar, O.; Athie, A.; Marchese, F.P.; González, J.; Bujanda, L.; Guo, S.; Matheu, A.; Huarte, M. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat. Commun. 2014, 5, 5812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dohner, H.; Fischer, K.; Bentz, M.; Hansen, K.; Benner, A.; Cabot, G.; Diehl, D.; Schlenk, R.; Coy, J.; Stilgenbauer, S. p53 gene deletion predicts for poor survival and non-response to therapy with purine analogs in chronic B-cell leukemias. Blood 1995, 85, 1580–1589. [Google Scholar] [PubMed]
- Ouillette, P.; Collins, R.; Shakhan, S.; Li, J.; Li, C.; Shedden, K.; Malek, S.N. The prognostic significance of various 13q14 deletions in chronic lymphocytic leukemia. Clin. Cancer Res. 2011, 17, 6778–6790. [Google Scholar] [CrossRef] [PubMed]
- Garding, A.; Bhattacharya, N.; Claus, R.; Ruppel, M.; Tschuch, C.; Filarsky, K.; Idler, I.; Zucknick, M.; Caudron-Herger, M.; Oakes, C.; et al. Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the In Cis Downregulation of a Gene Cluster That Targets NF-kB. PLoS Genet. 2013, 9, e1003373. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Q.; Wong, K.Y.; Li, Z.H.; Chim, C.S. Epigenetic silencing of tumor suppressor long non-coding RNA BM742401 in chronic lymphocytic leukemia. Oncotarget 2016, 7, 82400–82410. [Google Scholar] [CrossRef] [PubMed]
- Ronchetti, D.; Manzoni, M.; Agnelli, L.; Vinci, C.; Fabris, S.; Cutrona, G.; Matis, S.; Colombo, M.; Galletti, S.; Taiana, E.; et al. lncRNA profiling in early-stage chronic lymphocytic leukemia identifies transcriptional fingerprints with relevance in clinical outcome. Blood Cancer J. 2016, 6, e468. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, P.G.; Jares, P.; Rico, D.; Gómez-López, G.; Martínez-Trillos, A.; Villamor, N.; Ecker, S.; González-Pérez, A.; Knowles, D.G.; et al. Transcriptome characterization by RNA sequencing identifies a major molecular and clinical subdivision in chronic lymphocytic leukemia. Genome Res. 2014, 24, 212–226. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.R.; Ruppert, A.S.; Fobare, S.; Chen, T.L.; Liu, C.; Lehman, A.; Blachly, J.S.; Zhang, X.; Lucas, D.M.; Grever, M.R.; et al. The long noncoding RNA, treRNA, decreases DNA damage and is associated with poor response to chemotherapy in chronic lymphocytic leukemia. Oncotarget 2017, 8, 25942–25954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riccardo, B.; Alejandro, R.; Tiziana, D.; Giancarlo, C.; Tycho, B.; Julio, D.; Armando, L.; Antonella, Z.; Michele, D.; Vanessa, B.; et al. Expression of the transcribed ultraconserved region 70 and the related long non-coding RNA AC092652.2-202 has prognostic value in Chronic Lymphocytic Leukaemia. Br. J. Haematol. 2018. [Google Scholar] [CrossRef]
- Mack, G.S. To selectivity and beyond. Nat. Biotechnol. 2010, 28, 1259–1266. [Google Scholar] [CrossRef] [PubMed]
- Miranda, T.B.; Cortez, C.C.; Yoo, C.B.; Liang, G.; Abe, M.; Kelly, T.K.; Marquez, V.E.; Jones, P.A. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol. Cancer Ther. 2009, 8, 1579–1588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiskus, W.; Rao, R.; Balusu, R.; Ganguly, S.; Tao, J.; Sotomayor, E.; Mudunuru, U.; Smith, J.E.; Hembruff, S.L.; Atadja, P.; et al. Superior Efficacy of a Combined Epigenetic Therapy against Human Mantle Cell Lymphoma Cells. Clin. Cancer Res. 2012, 18, 6227–6238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oki, Y.; Buglio, D.; Fanale, M.; Fayad, L.; Copeland, A.; Romaguera, J.; Kwak, L.W.; Pro, B.; de Castro Faria, S.; Neelapu, S.; et al. Phase I Study of Panobinostat plus Everolimus in Patients with Relapsed or Refractory Lymphoma. Clin. Cancer Res. 2013, 19, 6882–6890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visser, H.P.J.; Gunster, M.J.; Kluin-Nelemans, H.C.; Manders, E.M.M.; Raaphorst, F.M.; Meijer, C.J.L.M.; Willemze, R.; Otte, A.P. The Polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. Br. J. Haematol. 2001, 112, 950–958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kienle, D.; Katzenberger, T.; Ott, G.; Saupe, D.; Benner, A.; Kohlhammer, H.; Barth, T.F.E.; Höller, S.; Kalla, J.; Rosenwald, A.; et al. Quantitative Gene Expression Deregulation in Mantle-Cell Lymphoma: Correlation with Clinical and Biologic Factors. J. Clin. Oncol. 2007, 25, 2770–2777. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Gupta, S.K.; Troska, T.P.; Nair, A.; Gupta, M. Long non-coding RNA profile in mantle cell lymphoma identifies a functional lncRNA ROR1-AS1 associated with EZH2/PRC2 complex. Oncotarget 2017, 8, 80223–80234. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, L.; Mathur, R.; Braun, F.K.; Wise, J.F.; Berkova, Z.; Neelapu, S.; Kwak, L.W.; Samaniego, F. FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia 2014, 28, 2376–2387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, D.; Li, H.; Li, R.; Sun, J.; Guo, P.; Han, H.; Yang, Y.; Li, J. Novel insight into MALAT-1 in cancer: Therapeutic targets and clinical applications. Oncol. Lett. 2016, 11, 1621–1630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, V.; Shen, Z.; Chakraborty, A.; Giri, S.; Freier, S.M.; Wu, X.; Zhang, Y.; Gorospe, M.; Prasanth, S.G.; Lal, A.; et al. Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB. PLoS Genet. 2013, 9, e1003368. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Jiang, Y.; Du, W.; Fairchild, L.; Melnick, A.; Elemento, O. Transcriptome sequencing reveals thousands of novel long non-coding RNAs in B cell lymphoma. Genome Med. 2015, 7, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Cheng, L.; Shi, H.; Zhang, Z.; Zhao, H.; Wang, Z.; Zhou, M. A potential panel of six-long non-coding RNA signature to improve survival prediction of diffuse large-B-cell lymphoma. Sci. Rep. 2016, 6, 27842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, M.; Zhao, H.; Xu, W.; Bao, S.; Cheng, L.; Sun, J. Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma. Mol. Cancer 2017, 16, 16. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Pannunzio, N.R.; Greisman, H.A.; Casero, D.; Parekh, C.; Lieber, M.R. Convergent BCL6 and lncRNA promoters demarcate the major breakpoint region for BCL6 translocations. Blood 2015, 126, 1730–1731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhang, M.; Xu, H.; Wang, Y.; Li, Z.; Chang, Y.; Wang, X.; Fu, X.; Zhou, Z.; Yang, S.; et al. Discovery and validation of the tumor-suppressive function of long noncoding RNA PANDA in human diffuse large B-cell lymphoma through the inactivation of MAPK/ERK signaling pathway. Oncotarget 2017, 8, 72182–72196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, W.; Fan, H.; Wu, G.; Wu, J.; Feng, J. Upregulation of long noncoding RNA PEG10 associates with poor prognosis in diffuse large B cell lymphoma with facilitating tumorigenicity. Clin. Exp. Med. 2016, 16, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Fang, S.; Liu, Y.; Li, X.; Liao, S.; Chen, J.; Liu, J.; Zhao, L.; Li, H.; Zhou, W.; et al. The long non-coding RNA NONHSAG026900 predicts prognosis as a favorable biomarker in patients with diffuse large B-cell lymphoma. Oncotarget 2017, 8, 34374–34386. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Li, H.; Guo, Y.; Luo, Y.; Li, H.; Xu, Y.; Deng, J.; Sun, B. A pilot study of long noncoding RNA expression profiling by microarray in follicular lymphoma. Gene 2016, 577, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Taub, R.; Kirsch, I.; Morton, C.; Lenoir, G.; Swan, D.; Tronick, S.; Aaronson, S.; Leder, P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7837–7841. [Google Scholar] [CrossRef] [PubMed]
- Dalla-Favera, R.; Bregni, M.; Erikson, J.; Patterson, D.; Gallo, R.C.; Croce, C.M. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7824–7827. [Google Scholar] [CrossRef] [PubMed]
- Doose, G.; Haake, A.; Bernhart, S.H.; López, C.; Duggimpudi, S.; Wojciech, F.; Bergmann, A.K.; Borkhardt, A.; Burkhardt, B.; Claviez, A.; et al. MINCR is a MYC-induced lncRNA able to modulate MYC’s transcriptional network in Burkitt lymphoma cells. Proc. Natl. Acad. Sci. USA 2015, 112, E5261–E5270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkle, M.; van den Berg, A.; Tayari, M.; Sietzema, J.; Terpstra, M.; Kortman, G.; de Jong, D.; Visser, L.; Diepstra, A.; Kok, K.; et al. Long noncoding RNAs as a novel component of the Myc transcriptional network. FASEB J. 2015, 29, 2338–2346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronchetti, D.; Manzoni, M.; Todoerti, K.; Neri, A.; Agnelli, L. In Silico Characterization of miRNA and Long Non-Coding RNA Interplay in Multiple Myeloma. Genes 2016, 7, 107. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zhao, H.; Wang, Z.; Cheng, L.; Yang, L.; Shi, H.; Yang, H.; Sun, J. Identification and validation of potential prognostic lncRNA biomarkers for predicting survival in patients with multiple myeloma. J. Exp. Clin. Cancer Res. 2015, 34, 102. [Google Scholar] [CrossRef] [PubMed]
- Hu, A.-X.; Huang, Z.-Y.; Zhang, L.; Shen, J. Potential prognostic long non-coding RNA identification and their validation in predicting survival of patients with multiple myeloma. Tumor Biol. 2017, 39, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Feng, Y.; Chen, H.; Huang, L.; Wang, F.; Bai, J.; Yang, Y.; Wang, J.; Zhao, W.; Jia, Y.; et al. Focusing on long non-coding RNA dysregulation in newly diagnosed multiple myeloma. Life Sci. 2018, 196, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Samur, M.K.; Minvielle, S.; Gulla, A.; Fulciniti, M.; Cleynen, A.; Aktas Samur, A.; Szalat, R.; Shammas, M.; Magrangeas, F.; Tai, Y.-T.; et al. Long intergenic non-coding RNAs have an independent impact on survival in multiple myeloma. Leukemia 2018. [Google Scholar] [CrossRef] [PubMed]
- Wenzhuo, Z.; Xueping, G.; Sijun, Y.; Moli, H.; Wenyue, Z.; Ping, C.; Xiaohui, Z.; Jinxiang, F.; Jing, Q.; Bingzong, L. Upregulation of lncRNA MEG3 Promotes Osteogenic Differentiation of Mesenchymal Stem Cells from Multiple Myeloma Patients By Targeting BMP4 Transcription. Stem Cells 2015, 33, 1985–1997. [Google Scholar] [CrossRef]
- Benetatos, L.; Dasoula, A.; Hatzimichael, E.; Georgiou, I.; Syrrou, M.; Bourantas, K.L. Promoter Hypermethylation of the MEG3 (DLK1/MEG3) Imprinted Gene in Multiple Myeloma. Clin. Lymphoma Myeloma 2008, 8, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.Y.; Li, Z.; Zhang, X.; Leung, G.K.K.; Chan, G.C.-f.; Chim, C.S. Epigenetic silencing of a long non-coding RNA KIAA0495 in multiple myeloma. Mol. Cancer 2015, 14, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Zhan, F.; Barlogie, B.; Arzoumanian, V.; Huang, Y.; Williams, D.R.; Hollmig, K.; Pineda-Roman, M.; Tricot, G.; van Rhee, F.; Zangari, M.; et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood 2007, 109, 1692–1700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poi, M.J.; Li, J.; Sborov, D.W.; VanGundy, Z.; Cho, Y.K.; Lamprecht, M.; Pichiorri, F.; Phelps, M.A.; Hofmeister, C.C. Polymorphism in ANRIL is associated with relapse in patients with multiple myeloma after autologous stem cell transplant. Mol. Carcinog. 2017, 56, 1722–1732. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Chen, J.; Zhang, H.; Wang, X.; Yao, H.; Peng, Y.; Zhang, W. LncRNA OIP5-AS1 loss-induced microRNA-410 accumulation regulates cell proliferation and apoptosis by targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple myeloma. Cell Death Dis. 2017, 8, e2975. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, L.; Hu, N.; Zhao, H. Long non-coding RNA FEZF1-AS1 promotes cell growth in multiple myeloma via miR-610/Akt3 axis. Biomed. Pharmacother. 2018, 103, 1727–1732. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hu, N.; Wang, C.; Zhao, H.; Gu, Y. Long non-coding RNA CCAT1 promotes multiple myeloma progression by acting as a molecular sponge of miR-181a-5p to modulate HOXA1 expression. Cell Cycle 2018, 17, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Malek, E.; Kim, B.; Driscoll, J. Identification of Long Non-Coding RNAs Deregulated in Multiple Myeloma Cells Resistant to Proteasome Inhibitors. Genes 2016, 7, 84. [Google Scholar] [CrossRef] [PubMed]
- Brocke-Heidrich, K.; Kretzschmar, A.K.; Pfeifer, G.; Henze, C.; Löffler, D.; Koczan, D.; Thiesen, H.-J.; Burger, R.; Gramatzki, M.; Horn, F. Interleukin-6—Dependent gene expression profiles in multiple myeloma INA-6 cells reveal a Bcl-2 family–independent survival pathway closely associated with Stat3 activation. Blood 2004, 103, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Binder, S.; Hösler, N.; Riedel, D.; Zipfel, I.; Buschmann, T.; Kämpf, C.; Reiche, K.; Burger, R.; Gramatzki, M.; Hackermüller, J.; et al. STAT3-induced long noncoding RNAs in multiple myeloma cells display different properties in cancer. Sci. Rep. 2017, 7, 7976. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhang, Y.; Wu, X.; Guo, Y.; Shi, W.; Qi, J.; Cong, H.; Wang, X.; Wu, X.; Ju, S. Upregulated lncRNA-PCAT1 is closely related to clinical diagnosis of multiple myeloma as a predictive biomarker in serum. Cancer Biomark. 2017, 18, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Kjems, J.; Damgaard, C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013, 73, 5609–5612. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B. Improved circRNA Identification by Combining Prediction Algorithms. Front. Cell Dev. Biol. 2018, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Szabo, L.; Salzman, J. Detecting circular RNAs: Bioinformatic and experimental challenges. Nat. Rev. Genet. 2016, 17, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.F.; Zhang, L.J.; Tan, K.; Jing, Q. Application of droplet digital PCR in quantitative detection of the cell-free circulating circRNAs. Biotechnol. Biotechnol. Equip. 2018, 32, 116–123. [Google Scholar] [CrossRef]
- Nicolet, B.P.; Engels, S.; Aglialoro, F.; van den Akker, E.; von Lindern, M.M.; Wolkers, M.C. Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. bioRxiv 2018. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Wang, S.; Ye, B.; Du, Y.; Li, C.; Xiong, Z.; Qu, Y.; Fan, Z. A Circular RNA Protects Dormant Hematopoietic Stem Cells from DNA Sensor cGAS-Mediated Exhaustion. Immunity 2018, 48, 688–701. [Google Scholar] [CrossRef] [PubMed]
- Guarnerio, J.; Bezzi, M.; Jeong, J.C.; Paffenholz, S.V.; Berry, K.; Naldini, M.M.; Lo-Coco, F.; Tay, Y.; Beck, A.H.; Pandolfi, P.P. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016, 165, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Dzikiewicz-Krawczyk, A.; Kok, K.; Slezak-Prochazka, I.; Robertus, J.-L.; Bruining, J.; Tayari, M.M.; Rutgers, B.; de Jong, D.; Koerts, J.; Seitz, A.; et al. ZDHHC11 and ZDHHC11B are critical novel components of the oncogenic MYC-miR-150-MYB network in Burkitt lymphoma. Leukemia 2017, 31, 1470–1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Du, W.W.; Wu, N.; Yang, W.; Awan, F.M.; Fang, L.; Ma, J.; Li, X.; Zeng, Y.; Yang, Z.; et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017, 24, 1609. [Google Scholar] [CrossRef] [PubMed]
- Dahl, M.; Daugaard, I.; Andersen, M.; Hansen, T.B.; Grønbæk, K.; Kjems, J.; Kristensen, L.S. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Lab. Investig. 2018. [Google Scholar] [CrossRef] [PubMed]
- Weng, W.; Wei, Q.; Toden, S.; Yoshida, K.; Nagasaka, T.; Fujiwara, T.; Cai, S.; Qin, H.; Ma, Y.; Goel, A. Circular RNA ciRS-7—A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin. Cancer Res. 2017, 23, 3918–3928. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, D.; Condorelli, A.; Ragusa, M.; Salito, L.; Sammito, M.; Banelli, B.; Caltabiano, R.; Barbagallo, G.; Zappalà, A.; Battaglia, R.; et al. Dysregulated miR-671-5p / CDR1-AS / CDR1 / VSNL1 axis is involved in glioblastoma multiforme. Oncotarget 2016, 7, 4746–4759. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okholm, T.L.H.; Nielsen, M.M.; Hamilton, M.P.; Christensen, L.-L.; Vang, S.; Hedegaard, J.; Hansen, T.B.; Kjems, J.; Dyrskjøt, L.; Pedersen, J.S. Circular RNA expression is abundant and correlated to aggressiveness in early-stage bladder cancer. NPJ Genom. Med. 2017, 2, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsiao, K.Y.; Lin, Y.C.; Gupta, S.K.; Chang, N.; Yen, L.; Sun, H.S.; Tsai, S.J. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017, 77, 2339–2350. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, D.; Caponnetto, A.; Cirnigliaro, M.; Brex, D.; Barbagallo, C.; D’Angeli, F.; Morrone, A.; Caltabiano, R.; Barbagallo, G.M.; Ragusa, M.; et al. CircSMARCA5 Inhibits Migration of Glioblastoma Multiforme Cells by Regulating a Molecular Axis Involving Splicing Factors SRSF1/SRSF3/PTB. Int. J. Mol. Sci. 2018, 19, 480. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Xu, Q.; Wang, Z.; Yang, Y.; Zhang, L.; Ma, J.; Sun, S.; Yang, F.; Zhou, W. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol. 2018, 68, 1214–1227. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Luo, J.; Hu, K.; Lin, J.; Huang, H.; Wang, Q.; Zhang, P.; Xiong, Z.; He, C.; Huang, Z.; et al. ZKSCAN1 gene and its related circular RNA (circ ZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol. Oncol. 2017, 11, 422–437. [Google Scholar] [CrossRef] [PubMed]
- Craig, V.J.; Cogliatti, S.B.; Imig, J.; Renner, C.; Neuenschwander, S.; Rehrauer, H.; Schlapbach, R.; Dirnhofer, S.; Tzankov, A.; Müller, A. Myc-mediated repression of microRNA-34a promotes high-grade transformation of B-cell lymphoma by dysregulation of FoxP1. Blood 2011, 117, 6227–6236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Yan, C.T.; Dou, Y.; Viboolsittiseri, S.S.; Wang, J.H. The role of a newly identified SET domain-containing protein, SETD3, in oncogenesis. Haematologica 2013, 98, 739–743. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; Johnson, N.A.; Severson, T.M.; Mungall, A.J.; An, J.; Goya, R.; Paul, J.E.; Boyle, M.; Woolcock, B.W.; Kuchenbauer, F. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010, 42, 181–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stilgenbauer, S.; Winkler, D.; Ott, G.; Schaffner, C.; Leupolt, E.; Bentz, M.; Möller, P.; Müller-Hermelink, H.K.; James, M.R.; Lichter, P.; et al. Molecular characterization of 11q deletions points to a pathogenic role of the ATM gene in mantle cell lymphoma. Blood 1999, 94, 3262–3264. [Google Scholar] [PubMed]
- Camus, V.; Miloudi, H.; Taly, A.; Sola, B.; Jardin, F. XPO1 in B cell hematological malignancies: From recurrent somatic mutations to targeted therapy. J. Hematol. Oncol. 2017, 10, 47. [Google Scholar] [CrossRef] [PubMed]
- Ahsmann, E.J.; Lokhorst, H.M.; Dekker, A.W.; Bloem, A.C. Lymphocyte function-associated antigen-1 expression on plasma cells correlates with tumor growth in multiple myeloma. Blood 1992, 79, 2068–2075. [Google Scholar] [PubMed]
- Chesi, M.; Nardini, E.; Lim, R.S.; Smith, K.D.; Kuehl, W.M.; Bergsagel, P.L. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 1998, 92, 3025–3034. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Middleton, R.E.; Sun, H.; Naniong, M.V.; Ott, C.J.; Mitsiades, C.S.; Wong, K.K.; Bradner, J.E.; Kaelin, W.G. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of ikaros proteins. Science 2014, 343, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Glazar, P.; Papavasileiou, P.; Rajewsky, N. circBase: A database for circular RNAs. RNA 2014, 20, 1666–1670. [Google Scholar] [CrossRef] [PubMed]
- Geiss, G.K.; Bumgarner, R.E.; Birditt, B.; Dahl, T.; Dowidar, N.; Dunaway, D.L.; Fell, H.P.; Ferree, S.; George, R.D.; Grogan, T.; et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008, 26, 317–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reis, P.P.; Waldron, L.; Goswami, R.S.; Xu, W.; Xuan, Y.; Perez-Ordonez, B.; Gullane, P.; Irish, J.; Jurisica, I.; Kamel-Reid, S. mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol. 2011, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Aird, D.; Ross, M.G.; Chen, W.-S.; Danielsson, M.; Fennell, T.; Russ, C.; Jaffe, D.B.; Nusbaum, C.; Gnirke, A. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011, 12, R18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kane, M.D.; Jatkoe, T.A.; Stumpf, C.R.; Lu, J.; Thomas, J.D.; Madore, S.J. Assessment of the sensitivity and specificity of oligonucleotide (50 mer) microarrays. Nucleic Acids Res. 2000, 28, 4552–4557. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I. Evolution to the rescue: Using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 2016, 17, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Lennox, K.A.; Behlke, M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016, 44, 863–877. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Baas, M.; Diederichs, S. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 2011, 21, 1944–1954. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goyal, A.; Myacheva, K.; Groß, M.; Klingenberg, M.; Duran Arqué, B.; Diederichs, S. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 2017, 45, e12. [Google Scholar] [CrossRef] [PubMed]
- Denzler, R.; McGeary, S.E.; Title, A.C.; Agarwal, V.; Bartel, D.P.; Stoffel, M. Impact of MicroRNA Levels, Target-Site Complementarity, and Cooperativity on Competing Endogenous RNA-Regulated Gene Expression. Mol. Cell 2016, 64, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Jens, M.; Rajewsky, N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat. Rev. Genet. 2014, 16, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Gardini, A. Global Run-On sequencing (GRO-seq). Methods Mol. Biol. 2017, 1468, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Dudekula, D.B.; Panda, A.C.; Grammatikakis, I.; De, S.; Abdelmohsen, K.; Gorospe, M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016, 13, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Jiang, S.; Kong, X.; El-Ali, N.; Ball Alexander R., J.; Ma, C.I.-H.; Hashimoto, N.; Yokomori, K.; Mortazavi, A. Single-nucleus RNA-seq of differentiating human myoblasts reveals the extent of fate heterogeneity. Nucleic Acids Res. 2016, 44, e158. [Google Scholar] [CrossRef] [PubMed]


| Name | Samples * | Expression | Proposed Function | Prognostic Impact | Reference |
|---|---|---|---|---|---|
| BALR-1 | 118 B-ALL | ↑ | No functional studies | No association with PFS/OS | [57] |
| 56 B-ALL | ↑ | No functional studies | No association with PFS/OS | [62] | |
| BALR-2 | 118 B-ALL, cell lines | ↑ | Promote cell survival via the inhibition of genes such as the proapoptotic BIM downstream of the glucocorticoid receptor | ↑ in steroid resistant patients and patients with poor OS | [57] |
| 56 B-ALL | ↑ | No functional studies | No association with PFS/OS | [62] | |
| BALR-6 | 118 B-ALL, cell lines, mice | ↑ | Promotes cell survival and inhibits apoptosis. Overexpression in mice leads to an increase in precursor cell populations | No association with PFS/OS | [57,58] |
| LINC00958 | 118 B-ALL | ↑ | No functional studies | No association with PFS/OS | [57] |
| 56 B-ALL | ↑ | No functional studies | No association with PFS/OS | [62] | |
| MALAT1 | 40 MCL, cell lines | ↑ | Binds to EZH2 and induces transcriptional repression of targets such as p21 and p27 | High vs. low expression: HR = 3 | [63] |
| DLBCL cell lines, xenograft mice | ↑ | KD induces the expression of autophagy-related proteins, improving chemotherapy-sensitivity | Not assessed | [64] | |
| 33 SMM, 170 MM, 36 PCL | ↑ | Associated with TP53-mediated DNA damage response | Not assessed | [65] | |
| 36 MM | ↑ | No functional studies | Change in expression (diagnosis /treatment) associated with PFS | [66] | |
| lincRNA-p21 | 73 primary CLL cells | ↑ in TP53wt compared to TP53del/mut | Decrease cell viability in a p53-dependent manner upon DNA damage | Not assessed | [67] |
| 68 CLL plasma samples | ↓ | p53 dependent cis-upregulation of p21, leading to cell cycle control through interaction with PRC2 | Not assessed | [12,68] | |
| NEAT1 | 73 primary CLL cells | ↑ in TP53wt compared to TP53del/mut | Nuclear retention of mRNAs with inverted repeats | Not assessed | [14,67] |
| 51 MM, cell lines | ↑ | Binds to miR-193a leading to MCL-1 upregulation and steroid resistance | ↑ in patients with poor OS | [69] | |
| MIAT | 67 CLL, cell lines | ↑ in patients with bad outcome | KD of MIAT, or its positive regulator OCT4, induces apoptosis | ↑ in patients with poor OS | [70] |
| 30 MM | ↓ in patients with t (11;14) | Involved in alternative splicing | [71,72] | ||
| CRNDE | 70 CLL | ↓ | Interacts with PRC2 and CoREST to modulate transcriptional repression | Promoter methylation associated with poor OS | [73,74] |
| 77 MM, cell lines | ↑ | Binds to miR-451 to induce proliferation and inhibit apoptosis | ↑ in patients with poor OS | [75] | |
| NAALADL2-AS2 | 10 DLBCL, cell lines | ↑ | Involved in p53, NFκB, and JAK/STAT signalling pathways (Gene Ontology Analysis) | Not assessed | [76,77] |
| HOTAIR | 50 DLBCL, cell lines | ↑ | Cell cycle progression and apoptosis inhibition through PI3K/AKT/NFκβ pathways | ↑ in patients with poor OS. HOTAIR > median vs. < median: HR = 3.1 | [78] |
| 164 DLBCL | Not specified | Recruits PRC2 and induce silencing of target genes through H3K27me3 | ↑ (higher than cancer-free tissue) in patients with favourable OS | [79,80] | |
| GAS5 | 33 SMM, 170 MM, 36 PCL | ↑ in patients with 1q gain lesions | No functional studies | Not assessed | [65] |
| MCL cell lines | KD reduces apoptosis and decreases the effects of mTOR inhibitors on cell viability | Not assessed | [81] | ||
| MCL cell lines | GAS5 interact with c-myc mRNA to reduce translation | Not assessed | [17] | ||
| UCA1 | 84 MM | ↓ | No functional studies | High vs. low expression: HR = 2 | [82] |
| 60 MM, cell lines | ↑ | Involved in cell proliferation and inhibition of apoptosis | Not assessed | [83] | |
| H19 | 30 MM | ↑ | Induce proliferation through NFκβ pathway | H19 ↑ in patients with poor PFS | [84] |
| 80 MM | ↑ | No functional studies | [85] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dahl, M.; Kristensen, L.S.; Grønbæk, K. Long Non-Coding RNAs Guide the Fine-Tuning of Gene Regulation in B-Cell Development and Malignancy. Int. J. Mol. Sci. 2018, 19, 2475. https://doi.org/10.3390/ijms19092475
Dahl M, Kristensen LS, Grønbæk K. Long Non-Coding RNAs Guide the Fine-Tuning of Gene Regulation in B-Cell Development and Malignancy. International Journal of Molecular Sciences. 2018; 19(9):2475. https://doi.org/10.3390/ijms19092475
Chicago/Turabian StyleDahl, Mette, Lasse Sommer Kristensen, and Kirsten Grønbæk. 2018. "Long Non-Coding RNAs Guide the Fine-Tuning of Gene Regulation in B-Cell Development and Malignancy" International Journal of Molecular Sciences 19, no. 9: 2475. https://doi.org/10.3390/ijms19092475