1. Introduction
Auxins are phytohormones of major importance for plant growth and development. There is extensive evidence of how this plant hormone regulates different physiological processes, such as cell development, including cell division, differentiation, and elongation, wall plasticity, tropisms, apical dominance, senescence, and flower development [
1,
2]. Indole-3-acetic acid (IAA) is the most common naturally occurring active auxin in plants. However, the active free IAA comprises only up to 25% of the total amount of IAA in a cell, depending on the tissue and plant species investigated. The majority of cellular auxin is found in inactive forms, conjugated to either amino acids, sugars, or small peptides [
3]. The metabolic control of the cellular auxin homeostasis through a tight regulation of de novo auxin biosynthesis, auxin degradation, and conjugation/deconjugation decisively orchestrates plant developmental processes [
4].
The role of auxin over the course of flower development has great importance in the formation of reproductive organs [
5]. For example, plants lacking the genes transport inhibitor response-1/auxin-binding F-box protein (
TIR/AFB), responsible for the auxin perception, present flowers with precocious pollen maturation [
6]. Additionally, mutant plants unable to express the auxin response factor 17 (
ARF17) gene display aberrant phenotypes at the pollen level, including changes in the cell-wall pattern affecting its viability [
7]. Also, two atypical members of the PIN auxin efflux carriers,
PIN5 and
PIN8, suggested as regulators of auxin homeostasis in pollen, participate in the pollen development and morphology [
8,
9]. At the same time, it was demonstrated that the complex interplay between auxin biosynthesis is of utmost importance for the development of floral organs, especially for anther dehiscence and pollen viability [
6]. Double mutants in the auxin biosynthesis-related
YUCCA (flavin monooxygenase-like enzymes) family genes
YUC2 and
YUC6, which are expressed during the early stages of anther and pollen development, present aberrant flower phenotypes, affecting stamina and anthers mainly. Accordingly, the
yuc2/yuc6 mutant plants are sterile [
10]. On the other hand, plants expressing the bacterial indole-3-acetic acid-lysine synthetase (
iaaL) gene driven by the
Arabidopsis thaliana phosphatidylinositol monophosphate 5-kinase 1 (
PIP5K1) promoter, which facilitates a tissue-specific expression in stamina filaments, showed shorter stamina filaments than those of wild-type plants, which is due to a reduced elongation of epidermal cells [
11]. The iaaL enzyme catalyzes the inactivation of free IAA through its conjugation to the amino acid
l-lysine. In addition, these plants also display a high rate of infertility. Pollen of these transgenic plants were unviable, because these pollen were unable to undergo the first round of pollen mitosis, generating aberrant pollen grain [
11]. Further indirect evidence for an accumulation of IAA in flowers was provided by employing the auxin signaling reporter construct DR5:β-glucuronidase (GUS) [
12]. The detailed histological analysis of the anthers at certain developmental stages determined the accumulation and dynamics of auxin in the tapetum, the middle layer, and the endothecium, which are the central tissues responsible for the dehiscence of the anther. An accumulation of auxin in the pollen during development was also observed [
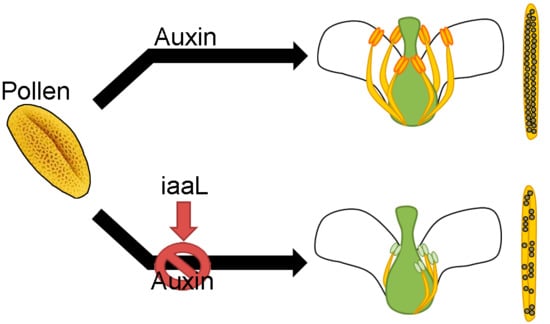
6]. Currently, the importance of auxin in floral development is demonstrated, but it is not known if the accumulation of auxin in the pollen affects the development of the flower. In this work, we demonstrate that the accumulation of auxin in the pollen grain plays an essential role for the stamina and anther development. Transgenic plants expressing the
iaaL gene driven by a pollen-specific promoter in the DR5:GUS genetic background were produced and analyzed. The corresponding plants were characterized as having significantly decreased free IAA levels in the pollen, presumably through the enzymatic conversion of IAA to indole-3-acetyl-
l-lysine (IAA-Lys). Consequently, the plants showed the loss of short stamina, as well as a loss of synchronization of stamina and anther development. Furthermore, they displayed defects in pollen tube development and had a smaller number of seeds. In summary, this work provides the first evidence for a role of auxin accumulation in the pollen grain, and how the deficiency of this hormone affects the proper development of the stamina and anthers.
3. Discussion
The role of auxin and its importance during floral development were previously extensively described [
4,
6,
10]. Nevertheless, to date, there are no studies connecting the accumulation of auxin in pollen grain and its potential role during development. The accumulation of auxin in pollen grain was previously described [
6], but whether or not this accumulation occurs during different stages of pollen development was never investigated. Using histological analysis of flower buds during different stages, we indirectly observed that there is a differential accumulation of auxin in pollen over the course of development, which is independent of the contribution of anther tissues that surround the pollen. The accumulation of auxin can be observed throughout the entire development of the pollen, from early to late stages (
Figure 1). We further investigated if this auxin accumulation in the male gametophyte plays a decisive role during flower development.
To decipher the role of auxin accumulation during pollen development, a genetic approach was chosen in which the levels of IAA were manipulated in a tissue-specific manner using the bacterial iaaL enzyme, which catalyzes the conjugation of IAA to
l-lysine, thereby generating an inactive form of IAA [
8]. Four promoters with different expression timing during pollen development were used to manipulate the expression of the
iaaL gene in the pollen of
Arabidopsis thaliana (
Figure S2). The selected promoters were designated as early pollen promoter (from the sugar transporter 2 gene,
STP2, At1g07340), intermediate pollen promoter 1 (from the sugar transporter 9 gene,
STP9, At1g50310), intermediate pollen promoter 2 (from the pollen-specific gene 2 gene,
PSG2, At1g28550), and late pollen promoter (from the phosphatase and tensin homolog deleted on chromosome 10 gene,
PTEN1, At5g39400) [
18,
19,
20,
21]. The resulting constructs were transformed into the DR5:GUS genetic background, to facilitate the analysis of IAA accumulation by monitoring the expression of the reporter gene
GUS under the control of the synthetic auxin-responsive DR5 promoter element [
12]. Using this strategy, we were able to reduce free IAA levels in the transgenic
pIPP2:
iaaL plants (
Figure 2). The feasibility of this strategy was previously proven [
11], expressing the
iaaL gene under the control of the promoter of the phosphatidylinositol monophosphate 5-kinase 1 (
PIP5K1) gene. This construct directs the expression of
iaaL gene to the stamina filament, decreasing the auxin content in stamina and consequently affecting the normal growth and development of these floral organs [
11]. The pollen grain-specific expression of the pollen-specific gene 2 (
PSG2) gene was previously reported as a pollen-specific promoter with exclusive activity in this floral organ [
20]. The described transcriptional properties of
PSG2 promised that the utilization of this promoter was suitable to trigger an increased conjugation rate of IAA in pollen grain through the induced activity of the gene product of the linked
iaaL gene, which, in turn, can be expected to reduce the amount of bio-active free IAA. Intriguingly, it was possible to detect a significant decrease in free IAA in flowers during an intermediate floral developmental stage (
Figure 3, stage II: 9–10) [
17], which is consistent with the spatiotemporal expression of the
pPSG2 (IPP2) promoter and also with the peak concentration of auxin in floral organs during development, as described previously [
22]. In contrast, no differences in free IAA were detected at early and late developmental stages (represented by stages I and III) [
17].
The reduced auxin levels in the transgenic
pIPP2:
iaaL plants, resulted in developmental defects in organs related to reproduction, such as the stamina and anthers (
Figure 4), pollen grains (
Figure 5), and seeds (
Figure 6). The
pIPP2:
iaaL plants exhibited problems in the development of short stamina. The proper anatomy of stamina (four long and two short stamina) is required to successfully complete the fertilization process in
Arabidopsis [
15,
16]. It was reported that a lack of an auxin transporter, such as
PIN6, produces defective short stamina or even the loss of these structures, a phenotype also observed in
pIPP2:
iaaL transgenic lines, suggesting that the reduction in auxin accumulated in pollen affects the development of the stamina [
23].
It is known that auxin is a pivotal plant hormone for proper control of plant development, including flower development and especially floral tissue maturation [
6]. IAA contributes to the regulation of the development and elongation of stamina, and the maturation of the anthers and pollen [
6]. For this reason, it can be concluded that the alteration of auxin levels in flowers is likely to affect dehiscence. The developments of anthers and pollen represent highly synchronized processes [
6,
15,
16]. Hence, by decreasing the amount of auxin in the pollen grain, it may be possible to affect the maturation and dehiscence of the anther, perturbing the synchronization of the linked processes and generating defects in anther breaking and pollen release. Transgenic
pIPP2:
iaaL plants, which show a decrease in the auxin levels, display perturbations in the synchronization of anther dehiscence (
Figure 7). The flowers of this transgenic line showed 41.5% of synchronization defects, including non-mature anthers, premature anthers, or anthers filled with water, which eventually do not lignify or break to release the pollen grain [
1,
5,
24], suggesting that an auxin reduction, particularly in pollen, affects the correct anther dehiscence.
The observed alterated phenotypes, such as a reduction in the number of stamina, seed content, and defective dehiscense in two independent
pIPP2:
iaaL transgenic lines, positively correlated with the reduction in auxin signaling as monitored by GUS activity assays (
Figure S5).
Furthermore, it is known that auxin is a driver of cell expansion, including pollen tube elongation [
4,
25]. Diminishing the amount of auxin in the pollen is supposed to affect the normal cylindrical shape of the pollen tube, resulting in aberrant balloon-shaped tips [
26,
27]. The
pIPP2:
iaaL pollen presented this phenotype in the tip of the pollen tube, suggesting that the reduction in auxin can affect the correct pollen tube formation (
Figure 5). Furthermore,
pIPP2:
iaaL plants also presented a lower number of seeds compared to their control, DR5:GUS (
Figure 6). This could be explained by the reduction in auxin present in the pollen, which ultimately affects the reproductive fitness of the plant similarly to the observation made in mutant plants lacking the
PIN8 gene, a pollen-specific auxin efflux carrier [
8,
9]. Therefore, it appears reasonable to conclude that the reduction in free auxin levels, specifically in pollen, can significantly affect the development of both the pollen grain and the elongation of the pollen tube, thereby affecting the reproduction and the fecundation rate of ovules, having an impact on the quantity of seeds in these transgenic plants.
The present work provides the first evidence for a central role of auxin accumulation in the pollen grain and its retrograde impact on stamina development via still unknown mechanisms which regulate the correct development of anthers and affect pollen germination and seed production.
4. Materials and Methods
4.1. Plant Growing
Arabidopsis thaliana ecotype Columbia (Col-0), DR5:GUS, and pPOLLEN:
iaaL seeds were surface-sterilized for 6 min with a solution of 50% (
v/
v) commercial bleach, and then washed with sterile distilled water. Seeds were germinated and grown on solid Murashige and Skoog (MS) medium (Phytotechnology Laboratories) [
28] containing 1% sucrose (
w/
v) and 0.8 (
w/
v) Phytagel (Sigma-Aldrich, St. Louis, MO, USA). After two weeks, plants were transferred to soil (Top Crop
TM Complete Mix, Santiago, Chile) and cultivated under a 16-h light/8-h dark cycle, at 23 °C. The pPOLLEN:
iaaL plant seeds were grown and selected in a medium supplemented with BASTA (glufosinate ammonium) at a final concentration of 30 μg/mL.
4.2. β-Glucuronidase (GUS) Assay and Histological Analysis
The GUS assay was performed according to Reference [
29]. Briefly, inflorescences were submerged in the GUS base solution buffer (100 mM NaH
2PO
4, pH 7.2; 10 mM EDTA, pH 8.0; 10% (
v/
v) methanol; 0.3% Triton X-100; 0.5 mM K
3Fe(CN)
6; 0.5 mM K
4Fe(CN)
6) supplemented with 1 mM 5-bromo-4-chloro-3-indolyl β-
d-glucuronide (X-Gluc). A vacuum of 40 mPa was applied for 15 min. Thereafter, the plant tissues were incubated for 12 h at 37 °C to facilitate the enzymatic conversion of the substrate 5-bromo-4-chloro-3-indolyl β-
d-glucuronide (X-gluc) to 5-bromo-4-chloro-3-indolyl, which was then oxidized and dimerized to give rise to the final insoluble reaction product (indigo). After the incubation, the tissues were washed with 70% ethanol until cleared. For the histological analysis, the GUS assay was performed and fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7) overnight. After these procedures, the samples were sent to the Service of Microscopy of the Universidad Catolica de Chile for the histological sections. The sections were analyzed using an optical microscope Leica model ICC50 HD (Leica, Wetzlar, Germany) and images were captured employing the computer program Leica Acquire (Leica, Wetzlar, Germany), with a magnification of 100× with immersion oil.
4.3. Statistical Analysis of Data
The data from the statistical analysis are represented as means with standard error (SE) (
Figure 3) and SD (
Figure 6). Statistical comparisons between groups was performed using a Student’s
t-test (
Figure 6), Fisher’s exact test (
Figure 4,
Figure 5 and
Figure 7), and two-way ANOVA/Bonferroni post test (
Figure 3) employing the Prism 6 software package (GraphPad Software, San Diego, California, CA, USA). Differences were considered statistically significant for
p < 0.05.
4.4. Cloning of Promoters and the iaaL Gene
The plasmid containing the
iaaL gene was kindly provided by Dr. Lars Østergaard of the John Innes Center, Norwich. The promoters were cloned from genomic DNA extracted from
Arabidopsis thaliana ecotype Columbia (Col-0). The DNA was extracted using the CTAB protocol [
30]. The primers to amplify the promoters were as follows (F—forward, R—reverse):
PAtSTP2 F: 5′-CACCAAACTCATTGCTTTCTCCTGA-3′;
PAtSTP2 R: 5′-TGTTGTTGATCTCTTAGCTTCT-3′;
PAtSTP9 F: 5′-CACCTGAGATTTAATGTGATGGT-3′;
PAtSTP9 R: 5′-TTATTTATTCTTCACTTATTGAT-3′;
PPSG2 F: 5′-CACCGGAAGAATACGAAGAAATAGTTGGC-3′;
pPSG2 R: 5′-CTTATTTCCGAAATAAACCTTTTTGC-3′;
PAtPTEN1 F: 5′-CACCGTGATCTGAGAAATGAGAGATTAC-3′;
PAtPTEN1 R: 5′-TCTGAAGGAAGAAAACATATCATTA-3′; iaaL F: 5′-CACCATGACTGCCTACGATAATGGA-3′; and iaaL R: 5′-TCAGTTTCGGCGGTCGAT-3′.
The PCR products were cloned into pENTRTM(Thermo-Fisher, Waltham, Massachusetts, MA, USA) according to the manufacturer’s instructions. For the multiple recombination of fragments included in entry vectors, the enzyme mixture LR clonaseTM II plus and the destination vector pB7m24GW,3 were used according to the manufacturer’s protocol.
4.5. Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
To perform the RT-PCR on RNA extracted from mature pollen grains, we used the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and cDNA was obtained using the SuperScript II First-Strand Synthesis System (Invitrogen). Pollen cDNA was used as a template for the RT-PCR. The amplification of target genes was achieved using a Phusion High-Fidelity DNA polymerase (Thermo-Fisher, Waltham, Massachusetts, MA, USA), according to the manufacturer’s instructions, performing 24 cycles. PCR products were then separated on Lafken brand 1% agarose gels. The primers used for RT-PCR were as follows: RT-GUS F: 5’-ATGTTACGTCCTGTAGAAACCCCAACC-3’; RT-GUS R: 5’-TGTTCGGCGTGGTGTAGAGCATTA-3’; RT-iaaL F 5’-CACCATGACTGCCTACGATAATGGA-3’; RT-iaaL R 5’-TCAGTTTCGGCGGTCGAT-3’; EF1α-1 F: 5’-TCACCCTTGGTGTCAAGCAGAT-3’; and EF1α-1 R: 5’-CAGGGTTGTATCCGACCTTCTT-3’.
4.6. Floral Organ Analysis
The optical inspection of stamina, anthers, and pollen was performed using a stereo microscope Leica model EZ4HD (Leica, Wetzlar, Germany). Images were captured through the software Leica Acquire (Leica, Wetzlar, Germany).
4.7. Alexander Stain
The Alexander stain was performed according to Reference [
31]. After the staining, the pollen was analyzed employing an optical microscope Leica model ICC50HD (Leica, Wetzlar, Germany) and photographs were taken using the software Leica Acquire (Leica, Wetzlar, Germany), with a magnification of 100× with immersion oil.
4.8. Pollen In Vitro Analysis
For in vitro germination tests of A. thaliana pollen grains, a solid germination medium containing 20% sucrose (w/v), 5 mM KCl, 5 mM CaCl2, 1 mM MgSO4, and 0.01% H3BO3 (w/v) was used. The pH of the medium was adjusted to 7.5 using a KOH solution, before being filtered. Then, low-melting agarose (Invitrogen) was added to a final concentration of 1.5% (w/v). After an incubation of 16 h in a humidity chamber, pollen tube growth was analyzed using optical microscopy Leica model ICC50HD (Leica, Wetzlar, Germany). Images were taken using the Leica Acquire software (Leica, Wetzlar, Germany).
4.9. Seed Analysis
Siliques of transgenic lines were collected and fixed in 100% ethanol. After the tissue lost its coloration, the siliques were inspected using optical microscopy Leica model EZ4HD (Leica, Wetzlar, Germany) and photos were taken using the Leica Acquire software (Leica, Wetzlar, Germany). Subsequently, the number of seeds in the inspected siliques was counted.
4.10. Auxin Analysis
Flowers from DR5:GUS and IPP2:
iaaL plants were classified according to four different floral developmental stages (I: 8–9, II: 10–11 and III: 12–13) [
17]. For each sample, flowers (40 mg) were harvested and shock-frozen in liquid nitrogen and stored at −80 °C. Auxin extraction was carried out as previously described [
32]. In brief, 1 mL of pre-warmed (65 °C) methanol was added to each sample. The extraction proceeded for another 60 min at room temperature under gentle shaking. Each sample was spiked with 50 pmol of (
2H
2)-IAA as a stable isotope-labeled internal standard. After centrifugation (1 min, 12,000 rpm), supernatants were transferred into fresh micro-reaction tubes and dried under vacuum. In order to pre-purify the samples for subsequent gas chromatography–tandem mass spectrometry analysis (GC–MS/MS), the dry extracts were dissolved in 50 µL of methanol and 200 µL of diethyl ether. Thereafter, they were loaded onto aminopropyl solid-phase extraction cartridges. Each cartridge was washed twice with 250 µL of CHCl
3:2-propanol (2:1,
v/
v). Next, the IAA-containing fraction was eluted with 400 µL of acidified diethyl ether (2% acetic acid,
v/
v). Thereafter, the eluates were transferred into 0.8-µL autosampler vials, and dried again in a gentle stream of nitrogen. Prior to mass spectrometric analysis, samples were derivatized by adding 20 µL of a mix consisting of 220 µL of acetone:methanol (9:1,
v/
v), 27 µL of diethyl ether, and 3 µL of a (trimethylsilyl)diazomethane solution (2.0 M in diethyl ether). To ensure complete derivatization, samples were incubated for 30 min at room temperature. Settings for the gas chromatograph and the mass spectrometer were as described previously [
32]. For IAA analysis, the following transitions were recorded: MeIAA,
m/
z 189 to
m/
z 130 (quantifier ion), and
m/
z 130 to
m/
z 103 (qualifier ion); (
2H
2)-MeIAA,
m/
z 191 to
m/
z 132 (quantifier ion), and
m/
z 132 to
m/
z 103 (qualifier ion). The measurements were done in triplicate and at least three samples for each floral stages was analyzed, before the amount of endogenous hormone content was calculated from the signal ratio of the unlabeled to the stable isotope-containing mass fragment observed in the parallel measurements.