Etoposide-Induced Apoptosis in Cancer Cells Can Be Reinforced by an Uncoupled Link between Hsp70 and Caspase-3
Abstract
:1. Introduction
2. Results
2.1. BT44 Inhibits Chaperone Activity of Hsp70 through Direct Binding to the Chaperone Molecule
2.1.1. Identification of Candidate Compounds
2.1.2. BT44 Inhibits the Chaperone Activity of Hsp70
2.1.3. BT44 Inhibits Hsp70 Activity in Cells
2.1.4. Inhibitory Actions of BT44 Occur throuh Direct Hsp70 Binding
2.2. BT44 Increases the Sensitivity of Cancer Cells with Over-Expressed Hsp70 to Etoposide-Induced Apoptosis
2.3. BT44 Enhances the Etoposide Sensitivity of U-937 Cells with High Hsp70 Levels
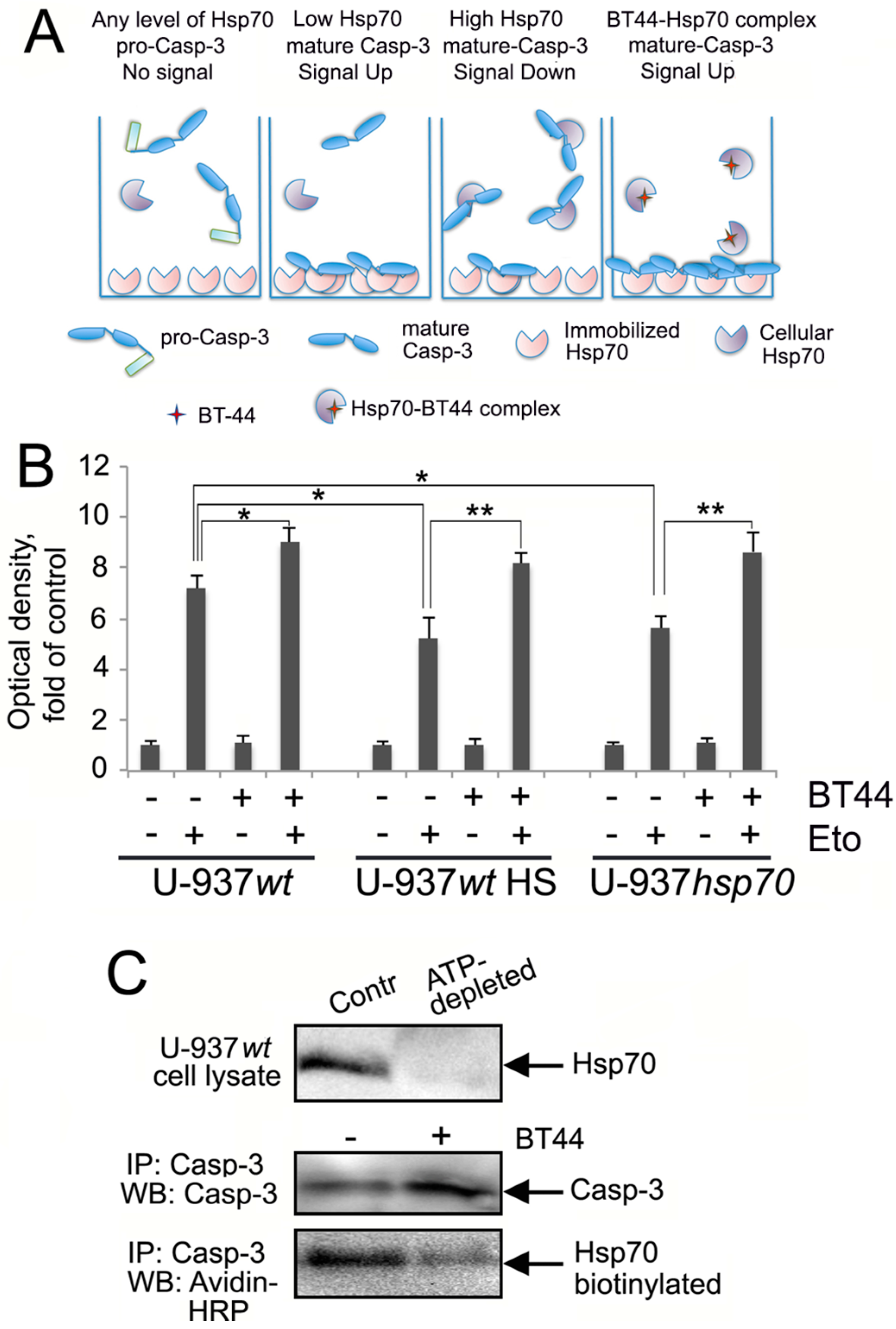
2.4. BT44 Prevents the Binding of Hsp70 to Caspase-3
3. Discussion
4. Materials and Methods
4.1. Virtual Screening of Compounds with Desirable Properties
4.2. Purification of Hsp70 and Measurement of Chaperone Activity
4.3. Cells
4.4. Refolding Assay
4.5. Drug Affinity Responsive Target Stability
4.6. Microscale Thermophoresis
4.7. Cytotoxicity Assay with xCELLigence System
4.8. Detection of Apoptosis
4.9. Western Blot Analysis
4.10. Caspase-3 Enzymatic Activity Assay
4.11. Competitive Protein–Protein Interaction Assay
4.12. Co-Immunoprecipitation
4.13. Statistics
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AIF | Apoptosis-Inducing Factor |
| APAF-1 | Apoptotic Protease Activating Factor 1 |
| CMLA | Carboxy-Methylated α-Lactalbumin |
| DARTS | Drug Affinity Responsive Target Stability |
| DTT | Dithiothreitol |
| EDTA | Ethylenediaminetetraacetic Acid |
| HRP | Horseradish Peroxidase |
| Hsp70 | Heat Shock Protein 70 kDa |
| IC50 | Half Maximal Inhibitory Concentration |
| JNK | c-Jun N-Terminal Kinase |
| MST | Microscale Thermophoresis |
| SAPK | Stress-Activated Protein Kinase |
| PAGE | Polyacrylamide Gel Electrophoresis |
| PASS | Prediction of Biological Activity Spectra for Substances |
| PVDF | Polyvinylidene Difluoride |
| TBS | Tris-Buffered Saline |
References
- Goloudina, A.R.; Demidov, O.N.; Garrido, C. Inhibition of Hsp70: A challenging anti-cancer strategy. Cancer Lett. 2012, 325, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R.; Evan, G.I. A matter of life and death. Cancer Cell 2002, 1, 19–30. [Google Scholar] [CrossRef]
- Mosser, D.D.; Morimoto, R.I. Molecular chaperones and the stress of oncogenesis. Oncogene 2004, 23, 2907–2918. [Google Scholar] [CrossRef] [PubMed]
- Ciocca, D.R.; Clark, G.M.; Tandon, A.K.; Fugua, S.A.; Welch, W.J.; McCuire, W.L. Heat shock protein Hsp70 in patient with axillary lymph node-negative breast cancer. Prognostic implication. J. Natl. Cancer Inst. 1993, 85, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Komarova, E.Y.; Afanasyeva, E.A.; Bulatova, M.M.; Cheetham, M.E.; Margulis, B.A.; Guzhova, I.V. Downstream caspases are novel targets for the antiapoptotic activity of the molecular chaperone Hsp70. Cell Stress Chaperones 2004, 9, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, S.K.; Gong, J. Heat shock proteins promote cancer: It’s a protection racket. Trends Biochem. Sci. 2016, 41, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Gabai, V.L.; Meriin, A.B.; Mosser, D.D.; Caron, A.W.; Rits, S.; Shifrin, V.I.; Sherman, M.Y. Hsp70 prevents activation of stress kinases. A novel pathway of cellular thermotolerance. J. Biol. Chem. 1997, 272, 18033–18037. [Google Scholar] [CrossRef] [PubMed]
- Meriin, A.B.; Yaglom, J.A.; Gabai, V.L.; Zon, L.; Ganiatsas, S.; Mosser, D.D.; Zon, L.; Sherman, M.Y. Protein-damaging stresses activate c-Jun N-terminal kinase via inhibition of its dephosphorylation: A novel pathway controlled by HSP72. Mol. Cell Biol. 1999, 19, 2547–2555. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, T.; Terada, K.; Oyadomari, S.; Mori, M. hsp70-DnaJ chaperone pair prevents nitric oxide- and CHOP-induced apoptosis by inhibiting translocation of Bax to mitochondria. Cell Death Differ. 2004, 11, 390–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saleh, A.; Srinivasula, S.M.; Balkir, L.; Robbins, P.D.; Alnemri, E.S. Negative regulation of the Apaf-1 apoptosome by Hsp70. Nat. Cell Biol. 2000, 2, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Beere, H.M.; Wolf, B.B.; Cain, K.; Mosser, D.D.; Mahboubi, A.; Kuwana, T.; Tailor, P.; Morimoto, R.I.; Cohen, G.M.; Green, D.R. Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat. Cell Biol. 2000, 2, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G. Heat shock protein 70 neutralizes apoptosis-inducing factor. Scientific World J. 2001, 1, 590–592. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Srinivasan, S.R.; Connarn, J.; Ahmad, A.; Young, Z.T.; Kabza, A.M.; Zuiderweg, E.R.; Sun, D.; Gestwicki, J.E. Analogs of the allosteric Heat shock protein 70 (Hsp70) Inhibitor, MKT-077, as anti-cancer agents. ACS Med. Chem. Lett. 2013, 4, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Hwang, I.; Lee, Y.S.; Park, S.; Lee, W.K.; Fernandes-Alnemri, T.; Alnemri, E.S.; Kim, Y.S.; Yu, J.W. Restoration of ASC expression sensitizes colorectal cancer cells to genotoxic stress-induced caspase-independent cell death. Cancer Lett. 2013, 331, 183–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stokes, J., III; Singh, U.P.; Scissum Gunn, K.; Acharya, A.; Manne, U.; Mishra, M. Targeting Hsp70: A possible therapy for cancer. Cancer Lett. 2016, 374, 156–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lomenick, B.; Olsen, R.W.; Huang, J. Identification of direct protein targets of small molecules. ACS Chem. Biol. 2011, 6, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Taldone, T.; Patel, H.J.; Bolaender, A.; Patel, M.R.; Chiosis, G. Protein chaperones: A composition of matter review (2008–2013). Expert Opin. Ther. Pat. 2014, 24, 501–518. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, R.; Sugihara, T.; Yoshida, A.; Nomura, H.; Reddel, R.R.; Simpson, R.; Maruta, H.; Kaul, S.C. Selective toxicity of MKT-077 to cancer cells is mediated by its binding to the Hsp70 family protein mot-2 and reactivation of p53 function. Cancer Res. 2000, 60, 6818–6821. [Google Scholar] [PubMed]
- Deocaris, C.C.; Widodo, N.; Ishii, T.; Kaul, S.C.; Wadhwa, R. Functional significance of minor structural and expression changes in stress chaperone mortalin. Ann. N. Y. Acad. Sci. 2007, 1119, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Leu, J.I.; Pimkina, J.; Frank, A.; Murphy, M.E.; George, D.L. A small molecule inhibitor of inducible heat shock protein 70. Mol. Cell 2009, 36, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Massey, A.J.; Williamson, D.S.; Browne, H.; Murray, J.B.; Dokurno, P.; Shaw, T.; Macias, A.T.; Daniels, Z.; Geoffroy, S.; Dopson, M.; et al. A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer Chemother. Pharmacol. 2010, 66, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Lazarev, V.F.; Sverchinsky, D.V.; Mikhaylova, E.R.; Semenyuk, P.I.; Komarova, E.Y.; Niskanen, S.A.; Nikotina, A.D.; Burakov, A.V.; Kartsev, V.G.; Guzhova, I.V.; et al. Sensitizing tumor cells to conventional drugs: Hsp70 chaperone inhibitors, their selection and application in cancer models. Cell Death Dis. 2018, 9, 41. [Google Scholar] [CrossRef] [PubMed]
- Nikotina, A.D.; Koludarova, L.; Komarova, E.Y.; Mikhaylova, E.R.; Aksenov, N.D.; Suezov, R.; Kartzev, V.G.; Margulis, B.A.; Guzhova, I.V. Discovery and optimization of cardenolides inhibiting HSF1 activation in human colon HCT-116 cancer cells. Oncotarget 2018, 9, 27268–27279. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciocca, D.R.; Calderwood, S.K. Heat shock proteins in cancer: Diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 2005, 10, 86–103. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Colvin, T.; Rauch, J.N.; Acosta-Alvear, D.; Kampmann, M.; Dunyak, B.; Hann, B.; Aftab, B.T.; Murnane, M.; Cho, M.; et al. Validation of the Hsp70-Bag3 protein-protein interaction as a potential therapeutic target in cancer. Mol. Cancer Ther. 2015, 14, 642–648. [Google Scholar] [CrossRef] [PubMed]
- Filimonov, D.A.; Lagunin, A.A.; Gloriozova, T.A.; Rudik, A.V.; Druzhilovskii, D.S.; Pogodin, P.V.; Poroikov, V.V. Prediction of the biological activity spectra of organic compounds using the PASS online web resource. Chem. Heterocycl. Compd. 2014, 50, 444–457. [Google Scholar] [CrossRef]
- Guzhova, I.V.; Lazarev, V.F.; Kaznacheeva, A.V.; Ippolitova, M.V.; Muronetz, V.I.; Kinev, A.V.; Margulis, B.A. Novel mechanism of Hsp70 chaperone-mediated prevention of polyglutamine aggregates in a cellular model of Huntington disease. Hum. Mol. Genet. 2011, 20, 3953–3963. [Google Scholar] [CrossRef] [PubMed]
- Wawrzynów, A.; Zylicz, M. Divergent effects of ATP on the binding of the DnaK and DnaJ chaperones to each other, or to their various native and denatured protein substrates. J. Biol. Chem. 1995, 270, 19300–19306. [Google Scholar] [CrossRef] [PubMed]
- Shevtsov, M.A.; Komarova, E.Y.; Meshalkina, D.A.; Bychkova, N.V.; Aksenov, N.D.; Abkin, S.V.; Margulis, B.A.; Guzhova, I.V. Exogenously delivered heat shock protein 70 displaces its endogenous analogue and sensitizes cancer cells to lymphocytes-mediated cytotoxicity. Oncotarget 2014, 5, 3101–3114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lomenick, B.; Hao, R.; Jonai, N.; Chin, R.M.; Aghajan, M.; Warburton, S.; Wang, J.; Wu, R.P.; Gomez, F.; Loo, J.A.; et al. Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. USA 2009, 106, 21984–21989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jerabek-Willemsen, M.; Wienken, C.J.; Braun, D.; Baaske, P.; Duhr, S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 2011, 9, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Thakur, M.; Mergel, K.; Weng, A.; Frech, S.; Gilabert-Oriol, R.; Bachran, D.; Melzig, M.F.; Fuchs, H. Real time monitoring of the cell viability during treatment with tumor-targeted toxins and saponins using impedance measurement. Biosens. Bioelectron. 2012, 35, 503–506. [Google Scholar] [CrossRef] [PubMed]





| Pa | Pi | Activity |
|---|---|---|
| 0.709 | 0.003 | Caspase 8 stimulant |
| 0.700 | 0.008 | Caspase 9 stimulant |
| 0.542 | 0.011 | Caspase 3 stimulant |
| 0.497 | 0.014 | Heat shock protein 27 antagonist |
| 0.465 | 0.022 | Transcription factor NF κ B stimulant |
| 0.378 | 0.029 | Heat shock protein antagonist |
| 0.387 | 0.082 | Transcription factor NF κ B1 inhibitor |
| 0.334 | 0.050 | Transcription factor NF κ B inhibitor |
| 0.301 | 0.048 | Heat shock protein 90 antagonist |
| Biological Activity | Number | IAP |
|---|---|---|
| Caspase 10 inhibitor | 6 | 0.996 |
| Caspase 2 inhibitor | 32 | 0.999 |
| Caspase 3 inhibitor | 1112 | 0.966 |
| Caspase 3 stimulant | 115 | 0.860 |
| Caspase 4 inhibitor | 9 | 0.885 |
| Caspase 6 inhibitor | 147 | 0.999 |
| Caspase 7 inhibitor | 937 | 0.971 |
| Caspase 8 inhibitor | 264 | 0.993 |
| Caspase 8 stimulant | 31 | 0.843 |
| Caspase 9 inhibitor | 44 | 0.968 |
| Caspase 9 stimulant | 39 | 0.779 |
| Caspase inhibitor | 7 | 0.999 |
| Caspase stimulant | 28 | 0.958 |
| Heat shock 70 kDa protein 1 antagonist | 16 | 0.924 |
| Heat shock factor protein 1 inhibitor | 271 | 0.855 |
| Heat shock protein 27 antagonist | 9 | 0.804 |
| Heat shock protein 70 agonist | 26 | 0.904 |
| Heat shock protein 70 antagonist | 23 | 0.965 |
| Heat shock protein 90 alpha antagonist | 1016 | 0.916 |
| Heat shock protein 90 antagonist | 1342 | 0.926 |
| Heat shock protein 90 β antagonist | 296 | 0.983 |
| Heat shock protein HSP 90 (HSC82) inhibitor | 11 | 0.999 |
| Heat shock protein agonist | 27 | 0.883 |
| Heat shock protein antagonist | 1374 | 0.923 |
| Transcription factor NF κ B inhibitor | 1009 | 0.910 |
| Transcription factor NF κ B stimulant | 7 | 0.882 |
| Transcription factor NF κ B1 inhibitor | 398 | 0.866 |
| Transcription factor NF κ B2 inhibitor | 74 | 0.990 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sverchinsky, D.V.; Nikotina, A.D.; Komarova, E.Y.; Mikhaylova, E.R.; Aksenov, N.D.; Lazarev, V.F.; Mitkevich, V.A.; Suezov, R.; Druzhilovskiy, D.S.; Poroikov, V.V.; et al. Etoposide-Induced Apoptosis in Cancer Cells Can Be Reinforced by an Uncoupled Link between Hsp70 and Caspase-3. Int. J. Mol. Sci. 2018, 19, 2519. https://doi.org/10.3390/ijms19092519
Sverchinsky DV, Nikotina AD, Komarova EY, Mikhaylova ER, Aksenov ND, Lazarev VF, Mitkevich VA, Suezov R, Druzhilovskiy DS, Poroikov VV, et al. Etoposide-Induced Apoptosis in Cancer Cells Can Be Reinforced by an Uncoupled Link between Hsp70 and Caspase-3. International Journal of Molecular Sciences. 2018; 19(9):2519. https://doi.org/10.3390/ijms19092519
Chicago/Turabian StyleSverchinsky, Dmitry V., Alina D. Nikotina, Elena Y. Komarova, Elena R. Mikhaylova, Nikolay D. Aksenov, Vladimir F. Lazarev, Vladimir A. Mitkevich, Roman Suezov, Dmitry S. Druzhilovskiy, Vladimir V. Poroikov, and et al. 2018. "Etoposide-Induced Apoptosis in Cancer Cells Can Be Reinforced by an Uncoupled Link between Hsp70 and Caspase-3" International Journal of Molecular Sciences 19, no. 9: 2519. https://doi.org/10.3390/ijms19092519