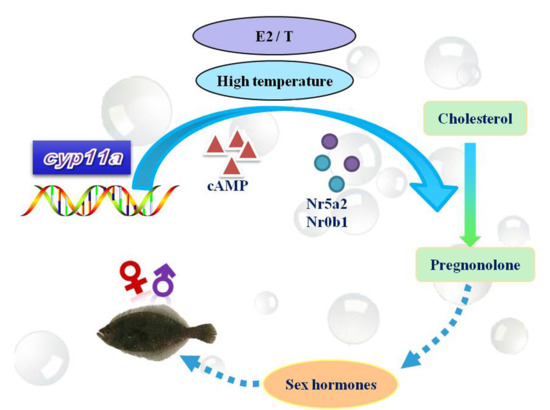
Characteristics of Cyp11a during Gonad Differentiation of the Olive Flounder Paralichthys olivaceus
Abstract
:1. Introduction
2. Results
2.1. Flounder cyp11a Gene and Its Phylogenetic Tree
2.2. Tissue Distribution of Flounder cyp11a
2.3. Spatial Expression Analysis of Flounder cyp11a Gene in Gonads
2.4. Analyses of cyp11a Expression, and Cholesterol and Pregnenolone Levels during Flounder Gonad Differentiation
2.4.1. Expression of Flounder cyp11a
2.4.2. Cholesterol and Pregnenolone Levels
2.5. Regulators’ Effect on Flounder cyp11a Expression
2.5.1. Impact of cAMP on Flounder cyp11a Expression in the Testis Cells
2.5.2. NR5a2 and NR0b1 Regulating Expression Level of cyp11a
3. Discussion
3.1. Characteristics of the Flounder cyp11a
3.2. Differential Patterns of cyp11a Expression, and Cholesterol and Pregnenolone Levels during the Flounder Gonad Differentiation
3.3. The Function of cAMP and Transcription Factors on Flounder cyp11a
4. Materials and Methods
4.1. Ethics Statement
4.2. Fish and Samples Collection
4.3. Total RNA Extraction, cDNA Synthesis and Genomic DNA Extraction
4.4. Cyp11a Cloning
4.5. Phylogenetic Tree Construction and Multiple Alignments
4.6. Semi-Quantitative Reverse Transcription Polymerase Chain Reaction (RT-PCR)
4.7. In Situ Hybridization (ISH) and Immunofluorescence Analyses
4.8. The Testis Cells Stimulated with Cyclic Adenosine Monophosphate (cAMP)
4.9. Cell Transfection Assay
4.10. Quantitative PCR (qPCR)
4.11. Enzyme-Linked Immunosorbent Assay (ELISA)
4.12. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Uno, T.; Ishizuka, M.; Itakura, T. Cytochrome P450 (CYP) in fish. Environ. Toxicol. Pharmacol. 2012, 34, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Guo, I.C.; Hu, M.C.; Chung, B.C. Transcriptional regulation of CYP11A1. J. Biomed. Sci. 2003, 10, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Tanaka, M.; Sakai, N.; Adachi, S.; Miller, W.L.; Nagahama, Y. Rainbow trout ovarian cholesterol side-chain cleavage cytochrome P450 (P450scc). cDNA cloning and mRNA expression during oogenesis. FEBS Lett. 1993, 319, 45–48. [Google Scholar] [CrossRef]
- Nelson, D.R. Comparison of P450s from human and fugu: 420 million years of vertebrate P450 evolution. Arch. Biochem. Biophys. 2003, 409, 18–24. [Google Scholar] [CrossRef]
- Kazeto, Y.; Ijiri, S.; Adachi, S.; Yamauchi, K. Cloning and characterization of a cDNA encoding cholesterol side-chain cleavage cytochrome P450 (CYP11A1): Tissue-distribution and changes in the transcript abundance in ovarian tissue of Japanese eel, Anguilla japonica, during artificially induced sexual development. J. Steroid Biochem. Mol. Biol. 2006, 99, 121–128. [Google Scholar] [PubMed]
- Zhang, W.; Zhou, L.; Senthilkumaran, B.; Huang, B.; Sudhakumari, C.C.; Kobayashi, T.; Nagahama, Y.; Wang, D. Molecular cloning of two isoforms of 11beta-hydroxylase and their expressions in the Nile tilapia, Oreochromis niloticus. Gen. Comp. Endocrinol. 2010, 165, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, M.; Uchida, K.; Abe, N.; Nozaki, M. Molecular cloning of cytochrome P450 side-chain cleavage and changes in its mRNA expression during gonadal development of brown hagfish, Paramyxine atami. Gen. Comp. Endocrinol. 2015, 212, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.J.; Hsiao, P.; Kuo, M.W.; Chung, B.C. Expression of zebrafish cyp11a1 as a maternal transcript and in yolk syncytial layer. Gene Expr. Patterns 2002, 2, 219–222. [Google Scholar] [CrossRef]
- Rajakumar, A.; Senthilkumaran, B. Expression analysis of cyp11a1 during gonadal development, recrudescence and after hCG induction and sex steroid analog treatment in the catfish, Clarias batrachus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2014, 176, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Govoroun, M.; McMeel, O.M.; D’Cotta, H.; Ricordel, M.-J.; Smith, T.; Fostier, A.; Guiguen, Y. Steroid enzyme gene expressions during natural and androgen-induced gonadal differentiation in the rainbow trout, Oncorhynchus mykiss. J. Exp. Zool. 2001, 290, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Shih, M.C.M.; Chiu, Y.N.; Hu, M.C.; Guo, I.C.; Chung, B.C. Regulation of steroid production: Analysis of Cyp11a1 promoter. Mol. Cell. Endocrinol. 2011, 336, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Morohashi, K.I.; Honda, S.I.; Inomata, Y.; Handa, H.; Omura, T. A common trans-acting factor Ad4-binding protein, to the promoters of steroidogenic P-450s. J. Biol. Chem. 1992, 267, 17913–17919. [Google Scholar] [PubMed]
- Parker, K.L. The roles of steroidogenic factor 1 in endocrine development and function. Mol. Cell. Endocrinol. 1998, 145, 15–20. [Google Scholar] [CrossRef]
- Ito, M.; Yu, R.; Jameson, J.L. DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol. Cell. Biol. 1997, 17, 1476–1483. [Google Scholar] [CrossRef] [PubMed]
- Zazopoulos, E.; Lalli, E.; Stocco, D.M.; Sassone-Corsi, P. DNA binding and transcriptional repression by DAX-1 blocks steroidogenesis. Nature 1997, 390, 311–315. [Google Scholar] [PubMed]
- Guo, I.C.; Chung, B.C. Cell-type specificity of human CYP11A1 TATA box. J. Steroid Biochem. Mol. Biol. 1999, 69, 329–334. [Google Scholar] [CrossRef]
- Schroeder, A.L.; Ankley, G.T.; Habib, T.; Garcia-Reyero, N.; Escalon, B.L.; Jensen, K.M.; Kahl, M.D.; Durhan, E.J.; Makynen, E.A.; Cavallin, J.E.; et al. Rapid effects of the aromatase inhibitor fadrozole on steroid production and gene expression in the ovary of female fathead minnows (Pimephales promelas). Gen. Comp. Endocrinol. 2017, 252, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, E. Studies on sex-manipulation and production of cloned populations in hirame, Paralichthys olivaceus (Temminck et Schlegel). Aquaculture 1999, 173, 235–246. [Google Scholar] [CrossRef]
- Shao, C.; Bao, B.; Xie, Z.; Chen, X.; Li, B.; Jia, X.; Yao, Q.; Ortí, G.; Li, W.; Li, X.; et al. The genome and transcriptome of Japanese flounder provide insights into flatfish asymmetry. Nat. Genet. 2016, 49, 119–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ospina-Alvarez, N.; Piferrer, F. Temperature-dependent sex determination in fish revisited: Prevalence, a single sex ratio response pattern, and possible effects of climate change. PLoS ONE 2008, 3, e2837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, P.; You, F.; Liu, M.; Wu, Z.; Wen, A.; Li, J.; Xu, Y.; Zhang, P. Steroid sex hormone dynamics during estradiol-17β induced gonadal differentiation in Paralichthys olivaceus (Teleostei). Chin. J. Oceanol. Limnol. 2010, 28, 254–259. [Google Scholar] [CrossRef]
- Sun, P.; You, F.; Ma, D.; Li, J.; Zhang, P. Sex steroid changes during temperature-induced gonadal differentiation in Paralichthys olivaceus (Temminck & Schegel, 1846). J. Appl. Ichthyol. 2013, 29, 886–890. [Google Scholar]
- Fan, Z.; You, F.; Wang, L.; Weng, S.; Wu, Z.; Hu, J.; Zou, Y.; Tan, X.; Zhang, P. Gonadal transcriptome analysis of male and female olive flounder Paralichthys olivaceus. BioMed Res. Int. 2014, 2014, 291067. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Fan, Z.; Weng, S.; Jiao, S.; Wu, Z.; Zou, Y.; Tan, X.; Li, J.; Zhang, P.; You, F. Characterization and expression of StAR2a and StAR2b in the olive flounder Paralichthys olivaceus. Gene 2017, 626, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; You, F.; Zhang, L.; Wen, A.; Wu, Z.; Xu, D.; Li, J.; Xu, Y.; Zhang, P. Histological evaluation of gonadal differentiation in olive flounder (Paralichthys olivaceus). Mar. Sci. 2009, 33, 53–58. [Google Scholar]
- Parajes, S.; Griffin, A.; Taylor, A.E.; Rose, I.T.; Miguel-Escalada, I.; Hadzhiev, Y.; Arlt, W.; Shackleton, C.; Müller, F.; Krone, N. Redefining the initiation and maintenance of zebrafish interrenal steroidogenesis by characterizing the key enzyme Cyp11a2. Endocrinology 2013, 154, 2702–2711. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Nakamura, M.; Kajiura-Kobayashi, H.; Young, G.; Nagahama, Y. Immunolocalization of steroidogenic enzymes (P450scc, P450c17, P450arom, and 3β-HSD) in immature and mature testes of rainbow trout (Oncorhynchus mykiss). Cell Tissue Res. 1998, 292, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Ings, J.S.; Van Der Kraak, G.J. Characterization of the mRNA expression of StAR and steroidogenic enzymes in zebrafish ovarian follicles. Mol. Reprod. Dev. 2006, 73, 943–954. [Google Scholar] [CrossRef] [PubMed]
- Baron, D.; Fostier, A.; Breton, B.; Guiguen, Y. Androgen and estrogen treatments alter steady state messengers RNA (mRNA) levels of testicular steroidogenic enzymes in the rainbow trout, Oncorhynchus mykiss. Mol. Reprod. Dev. 2005, 71, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Govoroun, M.; McMeel, O.M.; Mecherouki, H.; Smith, T.J.; Guiguen, Y. 17β-estradiol treatment decreases steroidogenic enzyme messenger ribonucleic acid levels in the rainbow trout testis. Endocrinology 2001, 142, 1841–1848. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Jing, J.; Wu, J.; Ma, W.; Dawar, F.U.; Mei, J.; Gui, J.-F. Characterization and sexual dimorphic expression of Cytochrome P450 genes in the hypothalamic-pituitary-gonad axis of yellow catfish. Gen. Comp. Endocrinol. 2015, 216, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Zou, Y.; Jiao, S.; Tan, X.; Wu, Z.; Liang, D.; Zhang, P.; You, F. Significant association of cyp19a promoter methylation with environmental factors and gonadal differentiation in olive flounder Paralichthys olivaceus. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2017, 208, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Blasco, M.; Fernandino, J.I.; Guilgur, L.G.; Strüssmann, C.A.; Somoza, G.M.; Vizziano-Cantonnet, D. Molecular characterization of cyp11a1 and cyp11b1 and their gene expression profile in pejerrey (Odontesthes bonariensis) during early gonadal development. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2010, 156, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, H.; Wang, F.; Zhang, W.; Peng, G. Nr0b1 (DAX1) mutation in zebrafish causes female-to-male sex reversal through abnormal gonadal proliferation and differentiation. Mol. Cell. Endocrinol. 2016, 433, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, Y.; Swain, A.; Weber, T.J.; Hentges, K.E.; Zanaria, E.; Lalli, E.; Tamai, K.T.; Sassone-Corsi, P.; Lovell-Badge, R.; Camerino, G.; et al. Steroidogenic factor 1 and Dax-1 colocalize in multiple cell lineages: Potential links in endocrine development. Mol. Endocrinol. 1996, 10, 1261–1272. [Google Scholar] [PubMed]
- Xu, B.; Yang, W.-H.; Gerin, I.; Hu, C.-D.; Hammer, G.D.; Koenig, R.J. Dax-1 and steroid receptor RNA activator (SRA) function as transcriptional coactivators for steroidogenic factor 1 in steroidogenesis. Mol. Cell. Biol. 2009, 29, 1719–1734. [Google Scholar] [CrossRef] [PubMed]
- You, F.; Liu, J.; Wang, X.; Xu, Y.; Huang, R.; Zhang, P. Study on embryonic development and early growth of triploid and gynogenetic diploid left-eyed flounder, Paralichthys olivaceus (T. et S.). Chin. J. Oceanol. Limnol. 2001, 19, 147–151. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kobayashi, T.; Kajiura-Kobayashi, H.; Nagahama, Y. Induction of XY sex reversal by estrogen involves altered gene expression in a teleost, tilapia. Cytogenet. Genome Res. 2003, 101, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Zheng, Y.; You, F.; Wu, Z.; Zou, Y.; Zhang, P. Establishment and characterization of a testicular Sertoli cell line from olive flounder Paralichthys olivaceus. Chin. J. Oceanol. Limnol. 2016, 34, 1054–1063. [Google Scholar] [CrossRef]
- Kowalewski, M.P.; Gram, A.; Boos, A. The role of hypoxia and HIF1α in the regulation of STAR-mediated steroidogenesis in granulosa cells. Mol. Cell. Endocrinol. 2015, 401, 35–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, W.; Sun, L. Evaluation of housekeeping genes as references for quantitative real time RT-PCR analysis of gene expression in Japanese flounder Paralichthys olivaceus. Fish Shellfish Immunol. 2011, 30, 638–645. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]










© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, D.; Fan, Z.; Zou, Y.; Tan, X.; Wu, Z.; Jiao, S.; Li, J.; Zhang, P.; You, F. Characteristics of Cyp11a during Gonad Differentiation of the Olive Flounder Paralichthys olivaceus. Int. J. Mol. Sci. 2018, 19, 2641. https://doi.org/10.3390/ijms19092641
Liang D, Fan Z, Zou Y, Tan X, Wu Z, Jiao S, Li J, Zhang P, You F. Characteristics of Cyp11a during Gonad Differentiation of the Olive Flounder Paralichthys olivaceus. International Journal of Molecular Sciences. 2018; 19(9):2641. https://doi.org/10.3390/ijms19092641
Chicago/Turabian StyleLiang, Dongdong, Zhaofei Fan, Yuxia Zou, Xungang Tan, Zhihao Wu, Shuang Jiao, Jun Li, Peijun Zhang, and Feng You. 2018. "Characteristics of Cyp11a during Gonad Differentiation of the Olive Flounder Paralichthys olivaceus" International Journal of Molecular Sciences 19, no. 9: 2641. https://doi.org/10.3390/ijms19092641