Excellent Degradation Performance of a Versatile Phthalic Acid Esters-Degrading Bacterium and Catalytic Mechanism of Monoalkyl Phthalate Hydrolase
Abstract
:1. Introduction
2. Results and Discussion
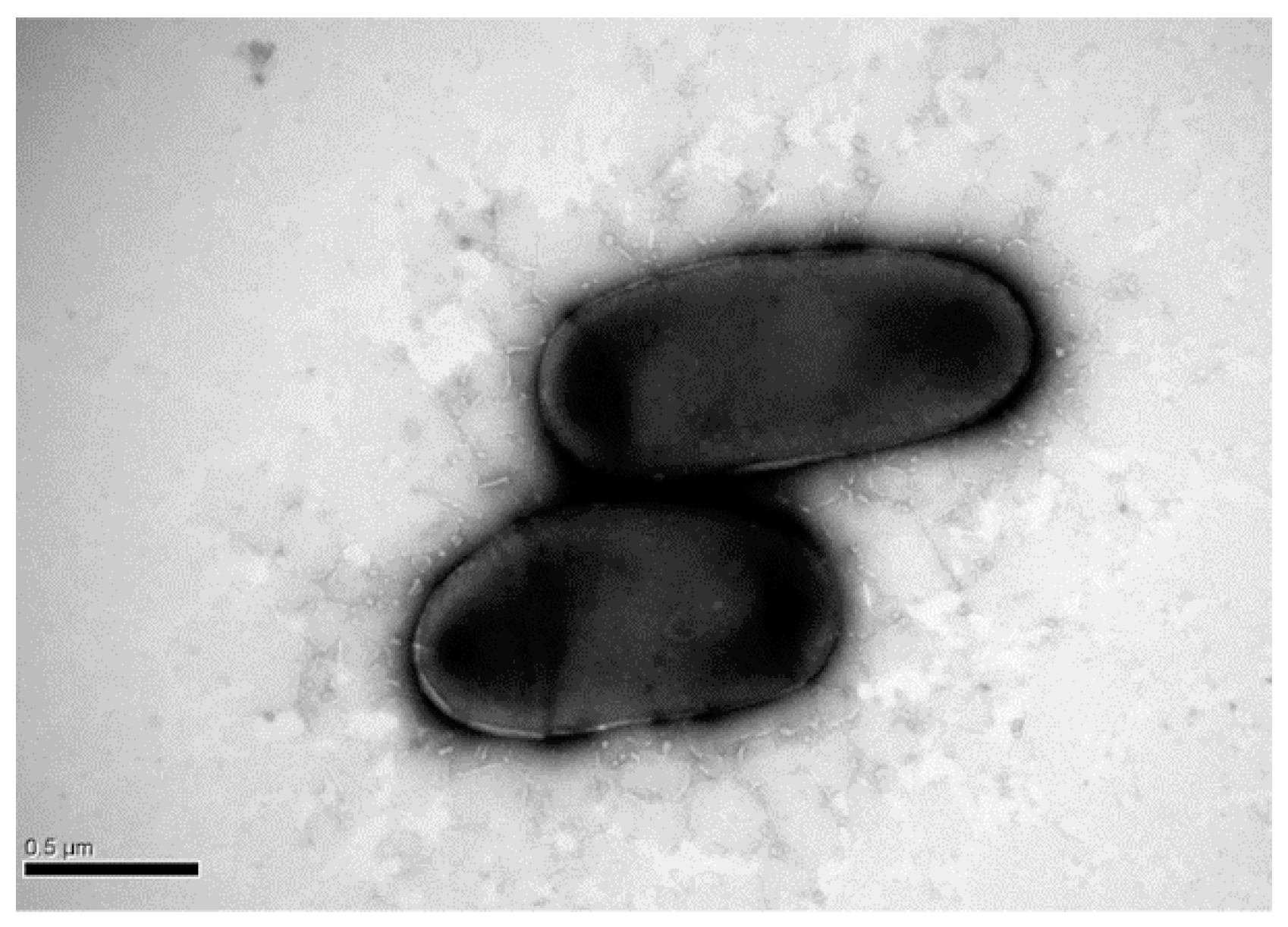
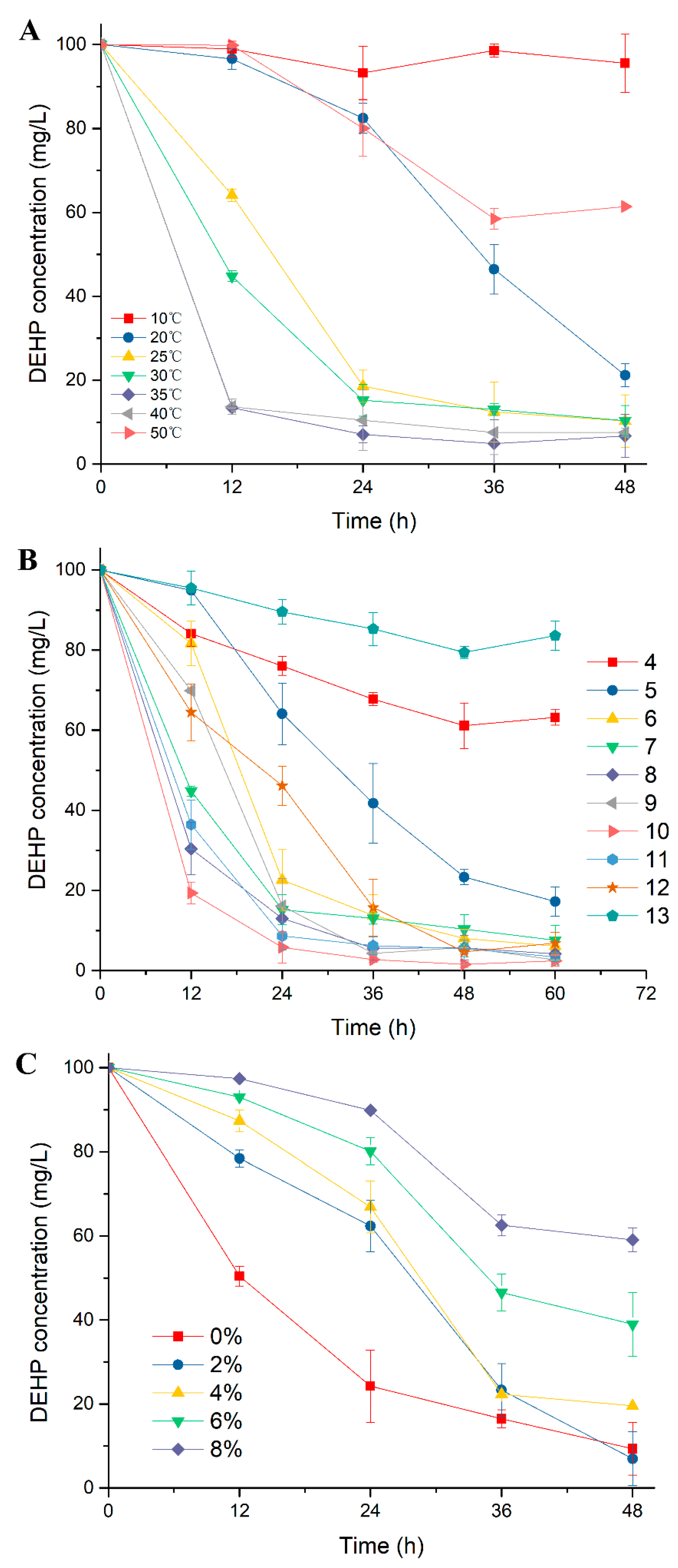
2.1. Excellent DEHP-Degrading Performance of Strain YC-JH1
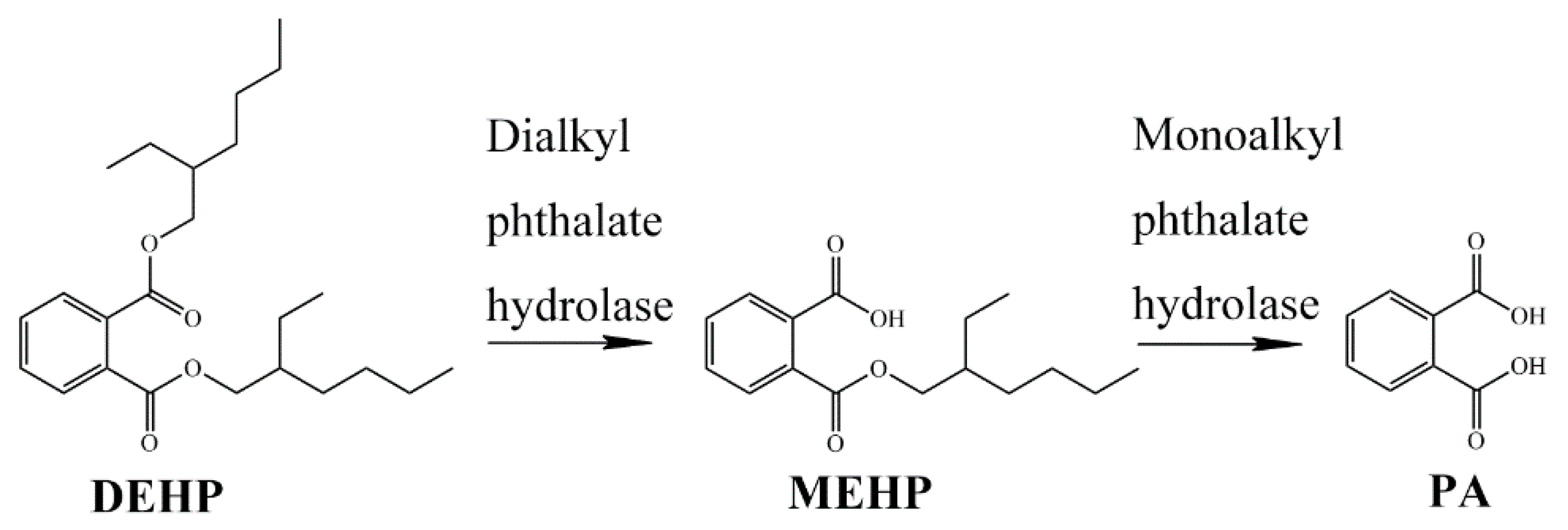
2.2. Catabolic Pathway of DEHP by Strain YC-JH1
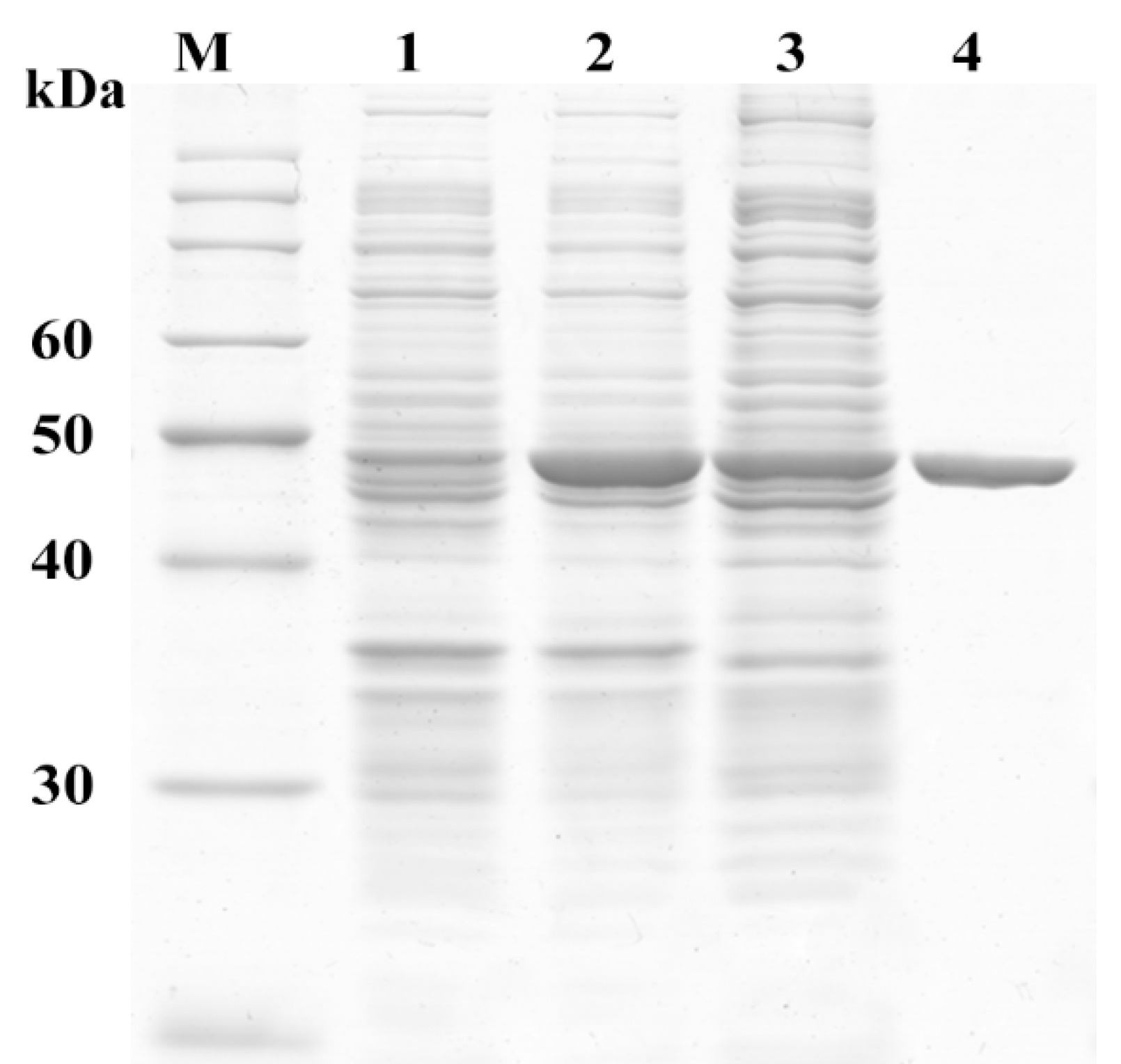
2.3. Sequence and Activity Analysis of MphG1
2.4. Interaction Mode of MphG1-MAP Complex
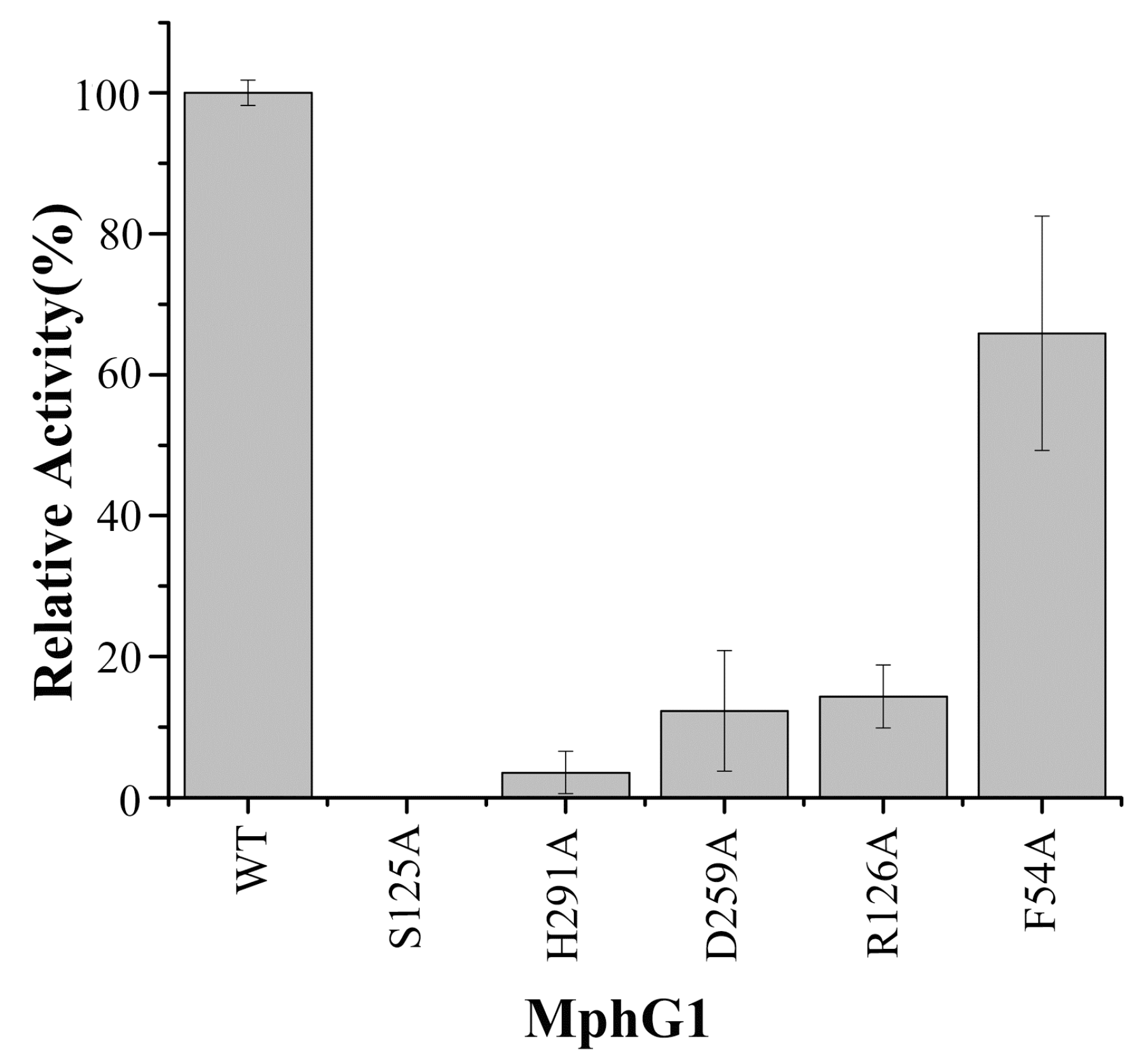
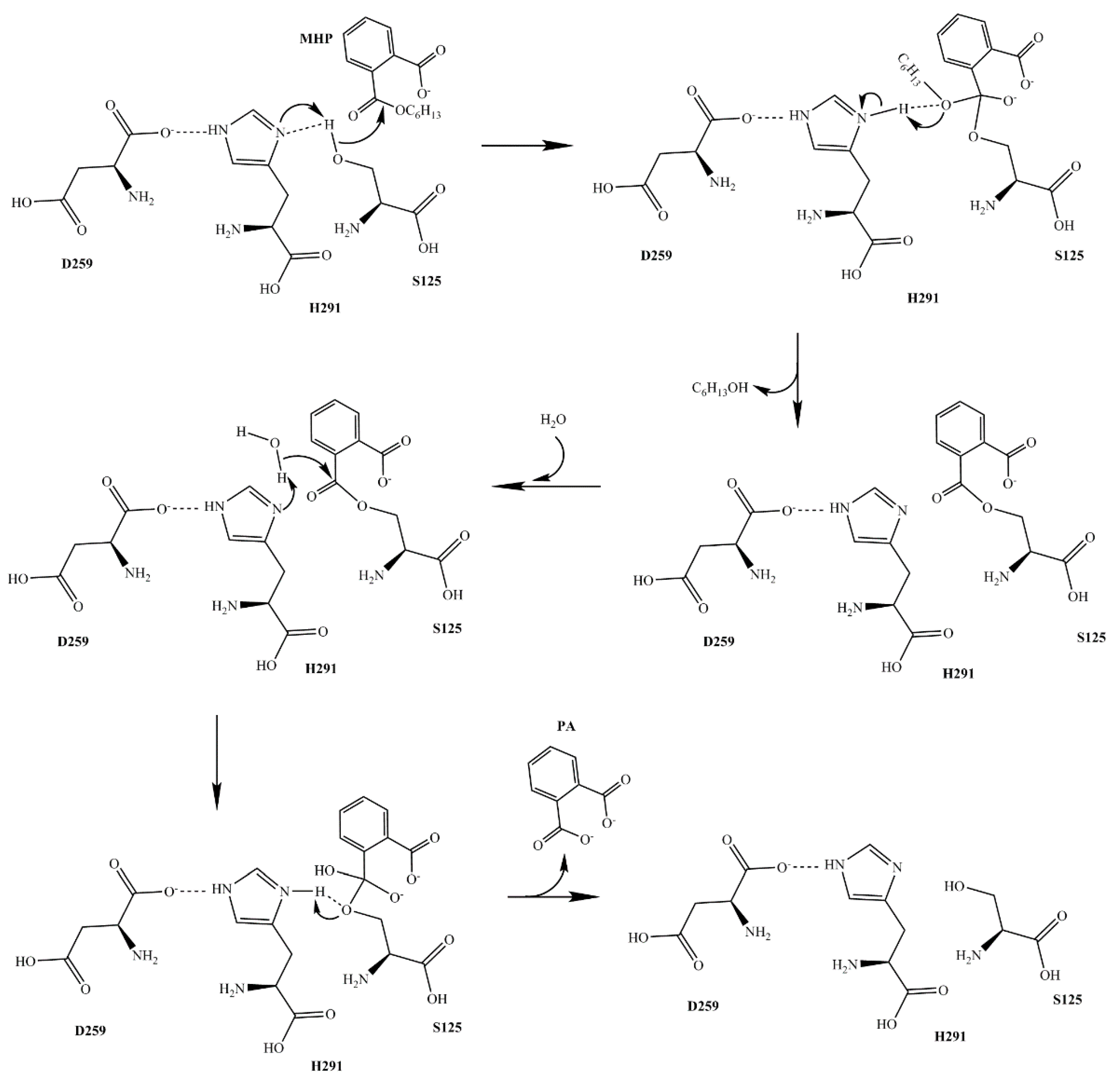
2.5. The Mutation Analysis and Catalytic Mechanism of MphG1
3. Materials and Methods
3.1. Chemicals, Medium and Strain
3.2. DEHP Degradation Assay by Strain YC-JH1
3.3. Identification of Intermediates of DEHP Degradation
3.4. Gene Cloning, Expression and Purification of Enzyme
3.5. Hydrolase Activity Assay
3.6. Molecular Docking and Molecular Dynamics Simulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gao, D.W.; Wen, Z.D. Phthalate esters in the environment: A critical review of their occurrence, biodegradation, and removal during wastewater treatment processes. Sci. Total Environ. 2016, 541, 986–1001. [Google Scholar] [CrossRef] [PubMed]
- Zolfaghari, M.; Drogui, P.; Seyhi, B.; Brar, S.K.; Buelna, G.; Dube, R. Occurrence, fate and effects of Di (2-ethylhexyl) Phthalate in wastewater treatment plants: A review. Environ. Pollut. 2014, 194, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Kamrin, M.A. Phthalate risks, phthalate regulation, and public health: A review. J. Toxicol. Environ. Health B Crit. Rev. 2009, 12, 157–174. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.; Hanaoka, T.; Yoshimura, M.; Zhang, S.; Wang, P.; Tsukino, H.; Inoue, K.; Nakazawa, H.; Tsugane, S.; Takahashi, K. Decreased serum free testosterone in workers exposed to high levels of di-n-butyl phthalate (DBP) and di-2-ethylhexyl phthalate (DEHP): A cross-sectional study in China. Environ. Health Perspect. 2006, 114, 1643–1648. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.S.; Chen, L.C.; Chen, B.S.; Lin, S.H. Bioremediation of endocrine disruptor di-n-butyl phthalate ester by Deinococcus radiodurans and Pseudomonas stutzeri. Chemosphere 2010, 78, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, C.; Zhu, Y.; Li, J.; Li, X. Biodegradation of seven phthalate esters by Bacillus mojavensis B1811. Int. Biodeter. Biodegr. 2018, 132, 200–207. [Google Scholar] [CrossRef]
- Wen, Z.; Gao, D.; Wu, W. Biodegradation and kinetic analysis of phthalates by an Arthrobacter strain isolated from constructed wetland soil. Appl. Microbiol. Biot. 2014, 98, 4683–4690. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Lu, Q.; de Toledo, R.A.; Shim, H. Degradation of di-2-ethylhexyl phthalate (DEHP) by an indigenous isolate Acinetobacter sp. SN13. Int. Biodeter. Biodegr. 2017, 117, 205–214. [Google Scholar] [CrossRef]
- Jin, D.; Kong, X.; Liu, H.; Wang, X.; Deng, Y.; Jia, M.; Yu, X. Characterization and Genomic Analysis of a Highly Efficient Dibutyl Phthalate-Degrading Bacterium Gordonia sp. Strain QH-12. Int. J. Mol. Sci. 2016, 17, 1012. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Niu, C.; Lu, Z. Individual or synchronous biodegradation of di-n-butyl phthalate and phenol by Rhodococcus ruber strain DP-2. J. Hazard. Mater. 2014, 273, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Zeng, P.; Moy, B.Y.; Song, Y.H.; Tay, J.H. Biodegradation of dimethyl phthalate by Sphingomonas sp. isolated from phthalic-acid-degrading aerobic granules. Appl. Microbiol. Biotechnol. 2008, 80, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Kong, X.; Cui, B.; Bai, Z.; Zhang, H. Biodegradation of di-n-butyl phthalate by a newly isolated halotolerant Sphingobium sp. Int. J. Mol. Sci. 2013, 14, 24046–24054. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Jia, Y.; Ruth, N.; Qiao, C.; Wang, J.; Zhao, B.; Yan, Y. Biodegradation of phthalic acid esters by a newly isolated Mycobacterium sp. YC-RL4 and the bioprocess with environmental samples. Environ. Sci. Pollut. Res. Int. 2016, 23, 16609–16619. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liang, R.; Dai, Q.; Jin, D.; Wang, Y.; Chao, W. Complete degradation of di-n-octyl phthalate by biochemical cooperation between Gordonia sp. strain JDC-2 and Arthrobacter sp. strain JDC-32 isolated from activated sludge. J. Hazard. Mater. 2010, 176, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.M.; Du, H.; Lin, J.; Chen, X.B.; Li, Y.W.; Li, H.; Cai, Q.Y.; Mo, C.H.; Qin, H.M.; Wong, M.H. Complete degradation of the endocrine disruptor di-(2-ethylhexyl) phthalate by a novel Agromyces sp. MT-O strain and its application to bioremediation of contaminated soil. Sci. Total Environ. 2016, 562, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, J.; Chowdhury, P.P.; Dutta, T.K. Complete degradation of di-n-octyl phthalate by Gordonia sp. strain Dop5. Chemosphere 2013, 90, 2571–2577. [Google Scholar] [CrossRef] [PubMed]
- Navacharoen, A.; Vangnai, A.S. Biodegradation of diethyl phthalate by an organic-solvent-tolerant Bacillus subtilis strain 3C3 and effect of phthalate ester coexistence. Int. Biodeter. Biodegr. 2011, 65, 818–826. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Y.; Liang, R.; Dai, Q.; Jin, D.; Chao, W. Biodegradation of an endocrine-disrupting chemical di-n-butyl phthalate by newly isolated Agrobacterium sp. and the biochemical pathway. Process. Biochem. 2011, 46, 1090–1094. [Google Scholar] [CrossRef]
- Nahurira, R.; Ren, L.; Song, J.; Jia, Y.; Wang, J.; Fan, S.; Wang, H.; Yan, Y. Degradation of Di(2-Ethylhexyl) Phthalate by a Novel Gordonia alkanivorans Strain YC-RL2. Curr. Microbiol. 2017, 74, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liao, X.; Yu, F.; Wei, Z.; Yang, L. Cloning of a dibutyl phthalate hydrolase gene from Acinetobacter sp. strain M673 and functional analysis of its expression product in Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 2483–2491. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Fan, X.; Qiu, Y.J.; Li, C.Y.; Xing, S.; Zheng, Y.T.; Xu, J.H. Newly identified thermostable esterase from Sulfobacillus acidophilus: Properties and performance in phthalate ester degradation. Appl. Environ. Microbiol. 2014, 80, 6870–6878. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, X.; Yang, Y.; Yue, D.; Xiao, L.; Yang, L. Biodegradation of an endocrine-disrupting chemical di-n-butyl phthalate by newly isolated Camelimonas sp. and enzymatic properties of its hydrolase. Biodegradation 2015, 26, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Chen, X.; Wang, X.; Liao, X.; Xiao, L.; Miao, A.; Wu, J.; Yang, L. Identification and characterization of a cold-active phthalate esters hydrolase by screening a metagenomic library derived from biofilms of a wastewater treatment plant. PLoS ONE 2013, 8, e75977. [Google Scholar] [CrossRef] [PubMed]
- Whangsuk, W.; Sungkeeree, P.; Nakasiri, M.; Thiengmag, S.; Mongkolsuk, S.; Loprasert, S. Two endocrine disrupting dibutyl phthalate degrading esterases and their compensatory gene expression in Sphingobium sp. SM42. Int. Biodeter. Biodegr. 2015, 99, 45–54. [Google Scholar] [CrossRef]
- Hong, D.K.; Jang, S.; Lee, C. Gene cloning and characterization of a psychrophilic phthalate esterase with organic solvent tolerance from an Arctic bacterium Sphingomonas glacialis PAMC 26605. J. Mol. Catal. B 2016, 133, S337–S345. [Google Scholar] [CrossRef]
- Nishioka, T.; Iwata, M.; Imaoka, T.; Mutoh, M.; Egashira, Y.; Nishiyama, T.; Shin, T.; Fujii, T. A mono-2-ethylhexyl phthalate hydrolase from a Gordonia sp. that is able to dissimilate di-2-ethylhexyl phthalate. Appl. Environ. Microbiol. 2006, 72, 2394–2399. [Google Scholar] [CrossRef] [PubMed]
- Iwata, M.; Imaoka, T.; Nishiyama, T.; Fujii, T. Re-characterization of mono-2-ethylhexyl phthalate hydrolase belonging to the serine hydrolase family. J. Biosci. Bioeng. 2016, 122, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Hara, H.; Stewart, G.R.; Mohn, W.W. Involvement of a novel ABC transporter and monoalkyl phthalate ester hydrolase in phthalate ester catabolism by Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 2010, 76, 1516–1523. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, Y.; Li, L.; Han, J. Probing Ligand-Binding Modes and Binding Mechanisms of Benzoxazole-Based Amide Inhibitors with Soluble Epoxide Hydrolase by Molecular Docking and Molecular Dynamics Simulation. J. Phys. Chem. B 2012, 116, 10219–10233. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.; Zeng, S.; Qin, X.; Sun, X.; Zhang, S.; Zhao, X.; Yu, Z.; Li, L. Molecular Docking and Site-directed Mutagenesis of a Bacillus thuringiensis Chitinase to Improve Chitinolytic, Synergistic Lepidopteran- larvicidal and Nematicidal Activities. Int. J. Biol. Sci. 2015, 11, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Yao, J.; Zhang, Q.; Yu, C.; Chen, P.; Liu, W.; Peng, D.; Choi, M.M.F. An integrated approach of bioassay and molecular docking to study the dihydroxylation mechanism of pyrene by naphthalene dioxygenase in Rhodococcus sp. ustb-1. Chemosphere 2015, 128, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Wang, J.; Li, K.; Yang, T.; Jia, Y.; Zhao, B.; Yan, Y. Complete genome sequence of Gordonia sp. YC-JH1, a bacterium efficiently degrading a wide range of phthalic acid esters. J. Biotechnol. 2018, 279, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.A.; Li, X.; Li, J.; Cao, J.; Qiu, Z.; Zhao, Q.; Xu, C.; Shu, W. Degradation of environmental endocrine disruptor di-2-ethylhexyl phthalate by a newly discovered bacterium, Microbacterium sp. strain CQ0110Y. Appl. Microbiol. Biotechnol. 2007, 74, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Bai, Z.; Chang, D.; Hoefel, D.; Jin, B.; Wang, P.; Wei, D.; Zhuang, G. Biodegradation of di-n-butyl phthalate by an isolated Gordonia sp. strain QH-11: Genetic identification and degradation kinetics. J. Hazard. Mater. 2012, 221–222, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Tang, F.; Wang, Y.; Zhao, J.; Zeng, X.; Luo, Q.; Wang, L. Biodegradation of dimethyl phthalate, diethyl phthalate and di-n-butyl phthalate by Rhodococcus sp. L4 isolated from activated sludge. J. Hazard. Mater. 2009, 168, 938–943. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Niu, G.; Yang, W.; Cao, X. Di(2-ethylhexyl) phthalate biodegradation and denitrification by a Pseudoxanthomonas sp. strain. Bioresour. Technol. 2015, 180, 356–359. [Google Scholar] [CrossRef] [PubMed]
- Pradeep, S.; Josh, M.K.; Binod, P.; Devi, R.S.; Balachandran, S.; Anderson, R.C.; Benjamin, S. Achromobacter denitrificans strain SP1 efficiently remediates di(2-ethylhexyl)phthalate. Ecotoxicol. Environ. Saf. 2015, 112, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Adelaja, O.; Keshavarz, T.; Kyazze, G. The effect of salinity, redox mediators and temperature on anaerobic biodegradation of petroleum hydrocarbons in microbial fuel cells. J. Hazard. Mater. 2015, 283, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Salgado, P.; Kiene, R.; Wiebe, W.; Magalhaes, C. Salinity as a regulator of DMSP degradation in Ruegeria pomeroyi DSS-3. J. Microbiol. 2014, 52, 948–954. [Google Scholar] [CrossRef] [PubMed]
- Kartal, B.; Koleva, M.; Arsov, R.; van der Star, W.; Jetten, M.S.; Strous, M. Adaptation of a freshwater anammox population to high salinity wastewater. J. Biotechnol. 2006, 126, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Dalal, V.; Mahto, J.K.; Kumar, P. Biodegradation of phthalic acid esters (PAEs) and in silico structural characterization of mono-2-ethylhexyl phthalate (MEHP) hydrolase on the basis of close structural homolog. J. Hazard. Mater. 2017, 338, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Feng, N.X.; Yu, J.; Mo, C.H.; Zhao, H.M.; Li, Y.W.; Wu, B.X.; Cai, Q.Y.; Li, H.; Zhou, D.M.; Wong, M.H. Biodegradation of di-n-butyl phthalate (DBP) by a novel endophytic Bacillus megaterium strain YJB3. Sci. Total Environ. 2018, 616–617, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Li, P.Y.; Yao, Q.Q.; Wang, P.; Zhang, Y.; Li, Y.; Zhang, Y.Q.; Hao, J.; Zhou, B.C.; Chen, X.L.; Shi, M.; et al. A Novel Subfamily Esterase with a Homoserine Transacetylase-like Fold but No Transferase Activity. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Botos, I.; Wlodawer, A. The expanding diversity of serine hydrolases. Curr. Opin. Struct. Biol. 2007, 17, 683–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajagopalan, S.; Wang, C.; Yu, K.; Kuzin, A.P.; Richter, F.; Lew, S.; Miklos, A.E.; Matthews, M.L.; Seetharaman, J.; Su, M.; et al. Design of activated serine-containing catalytic triads with atomic-level accuracy. Nat. Chem. Biol. 2014, 10, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Nardini, M.; Dijkstra, B.W. Alpha/beta hydrolase fold enzymes: The family keeps growing. Curr. Opin. Struct. Biol. 1999, 9, 732–737. [Google Scholar] [CrossRef]
- Dunn, G.; Montgomery, M.G.; Mohammed, F.; Coker, A.; Cooper, J.B.; Robertson, T.; Garcia, J.L.; Bugg, T.D.; Wood, S.P. The structure of the C-C bond hydrolase MhpC provides insights into its catalytic mechanism. J. Mol. Biol. 2005, 346, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Ruzzini, A.C.; Bhowmik, S.; Yam, K.C.; Ghosh, S.; Bolin, J.T.; Eltis, L.D. The lid domain of the MCP hydrolase DxnB2 contributes to the reactivity toward recalcitrant PCB metabolites. Biochemistry 2013, 52, 5685–5695. [Google Scholar] [CrossRef] [PubMed]
- Habe, H.; Morii, K.; Fushinobu, S.; Nam, J.W.; Ayabe, Y.; Yoshida, T.; Wakagi, T.; Yamane, H.; Nojiri, H.; Omori, T. Crystal structure of a histidine-tagged serine hydrolase involved in the carbazole degradation (CarC enzyme). Biochem. Biophys. Res. Commun. 2003, 303, 631–639. [Google Scholar] [CrossRef]
- Horsman, G.P.; Bhowmik, S.; Seah, S.Y.; Kumar, P.; Bolin, J.T.; Eltis, L.D. The tautomeric half-reaction of BphD, a C-C bond hydrolase. Kinetic and structural evidence supporting a key role for histidine 265 of the catalytic triad. J. Biol. Chem. 2007, 282, 19894–19904. [Google Scholar] [CrossRef] [PubMed]
- Ryan, A.; Polycarpou, E.; Lack, N.A.; Evangelopoulos, D.; Sieg, C.; Halman, A.; Bhakta, S.; Eleftheriadou, O.; McHugh, T.D.; Keany, S.; et al. Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach. Br. J. Pharmacol. 2017, 174, 2209–2224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
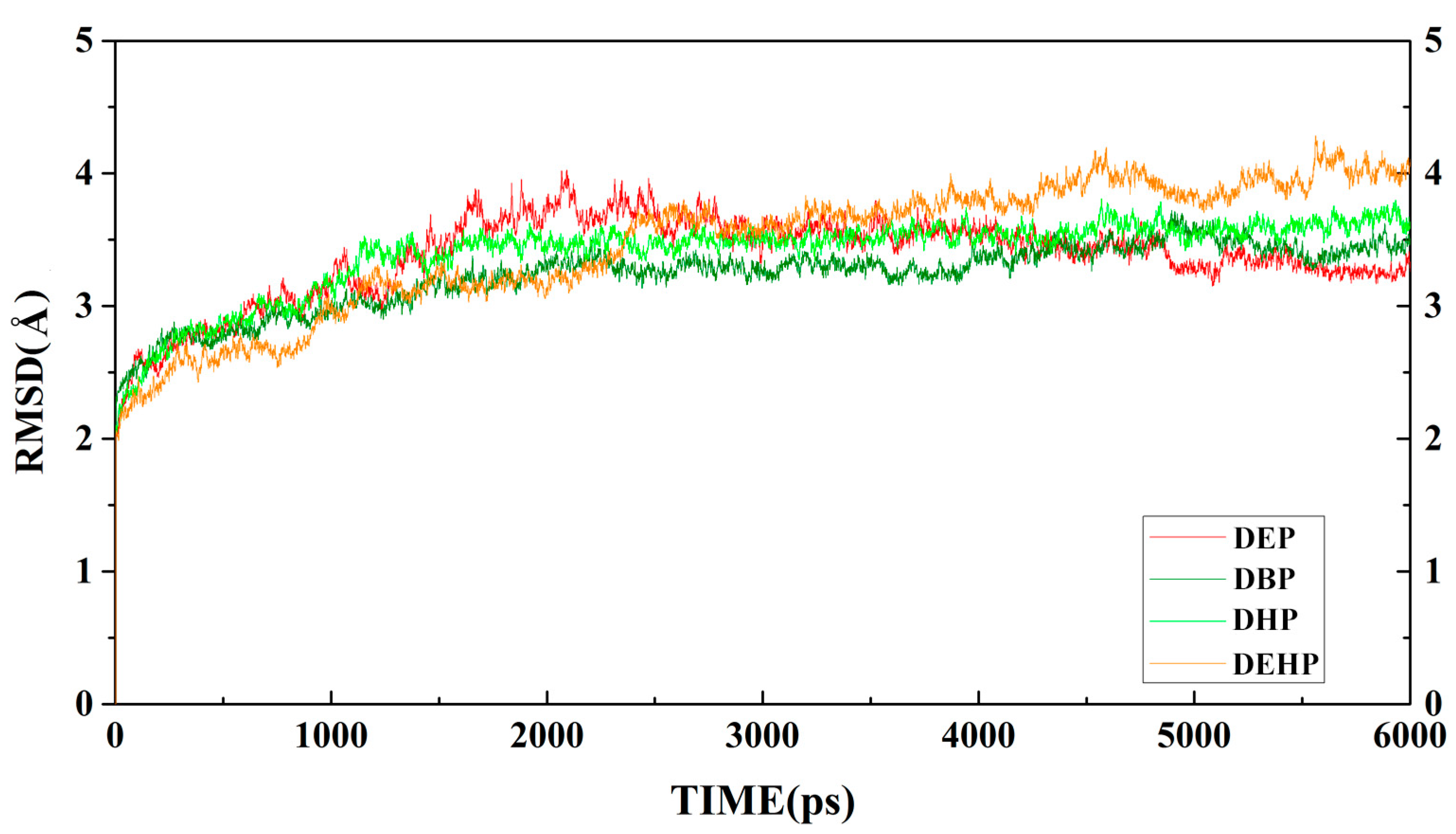

| Substrate | Structure | Binding Energy (kcal/mol) | Specific Activity (U/mg) |
|---|---|---|---|
| MEP | 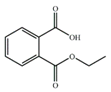 | −7.86 | 2.84 ± 0.14 |
| MBP |  | −9.20 | 2.99 ± 0.04 |
| MHP |  | −17.34 | 3.14 ± 0.06 |
| MEHP | 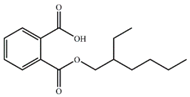 | −13.72 | 2.91 ± 0.09 |
| Complex | RMSD/Å |
|---|---|
| MphG1-MEP | 3.3774 ± 0.4 |
| MphG1-MBP | 3.2302 ± 0.4 |
| MphG1-MHP | 3.4007 ± 0.4 |
| MphG1-MEHP | 3.4492 ± 0.3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, S.; Wang, J.; Yan, Y.; Wang, J.; Jia, Y. Excellent Degradation Performance of a Versatile Phthalic Acid Esters-Degrading Bacterium and Catalytic Mechanism of Monoalkyl Phthalate Hydrolase. Int. J. Mol. Sci. 2018, 19, 2803. https://doi.org/10.3390/ijms19092803
Fan S, Wang J, Yan Y, Wang J, Jia Y. Excellent Degradation Performance of a Versatile Phthalic Acid Esters-Degrading Bacterium and Catalytic Mechanism of Monoalkyl Phthalate Hydrolase. International Journal of Molecular Sciences. 2018; 19(9):2803. https://doi.org/10.3390/ijms19092803
Chicago/Turabian StyleFan, Shuanghu, Junhuan Wang, Yanchun Yan, Jiayi Wang, and Yang Jia. 2018. "Excellent Degradation Performance of a Versatile Phthalic Acid Esters-Degrading Bacterium and Catalytic Mechanism of Monoalkyl Phthalate Hydrolase" International Journal of Molecular Sciences 19, no. 9: 2803. https://doi.org/10.3390/ijms19092803





