Genome-Wide Identification, Molecular Evolution, and Expression Divergence of Aluminum-Activated Malate Transporters in Apples
Abstract
:1. Introduction
2. Results
2.1. Identification and Characterization of ALMT Gene Family in Apples
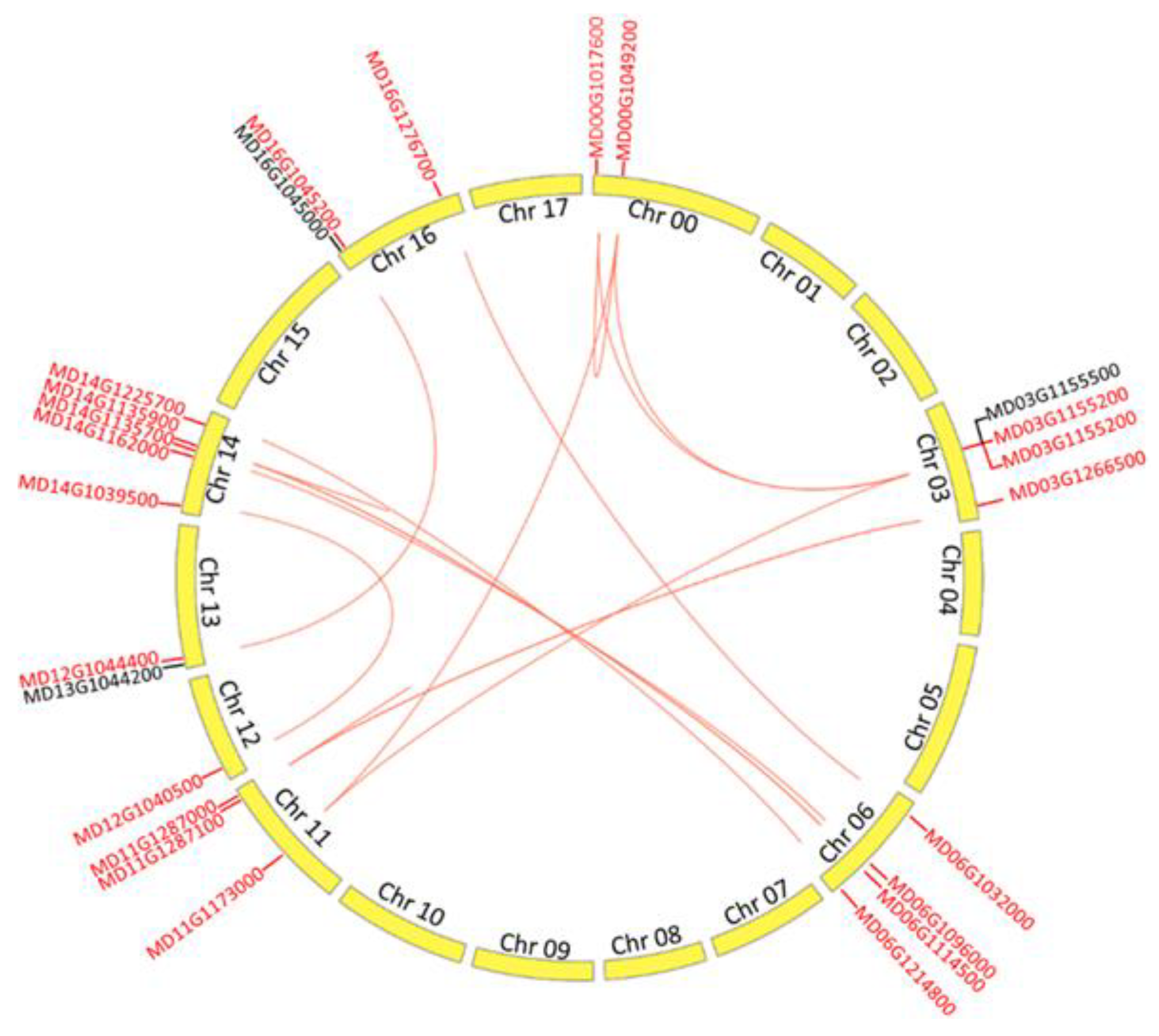
2.2. Chromosome Distribution and Duplication of MdALMT Genes
2.3. Phylogenetic Analysis, Gene Duplication, and Gene Loss in Four Rosaceae Species
2.4. Positive Selection Analysis of ALMT Genes in Four Rosaceae Species
2.5. Estimation of Functional Divergence
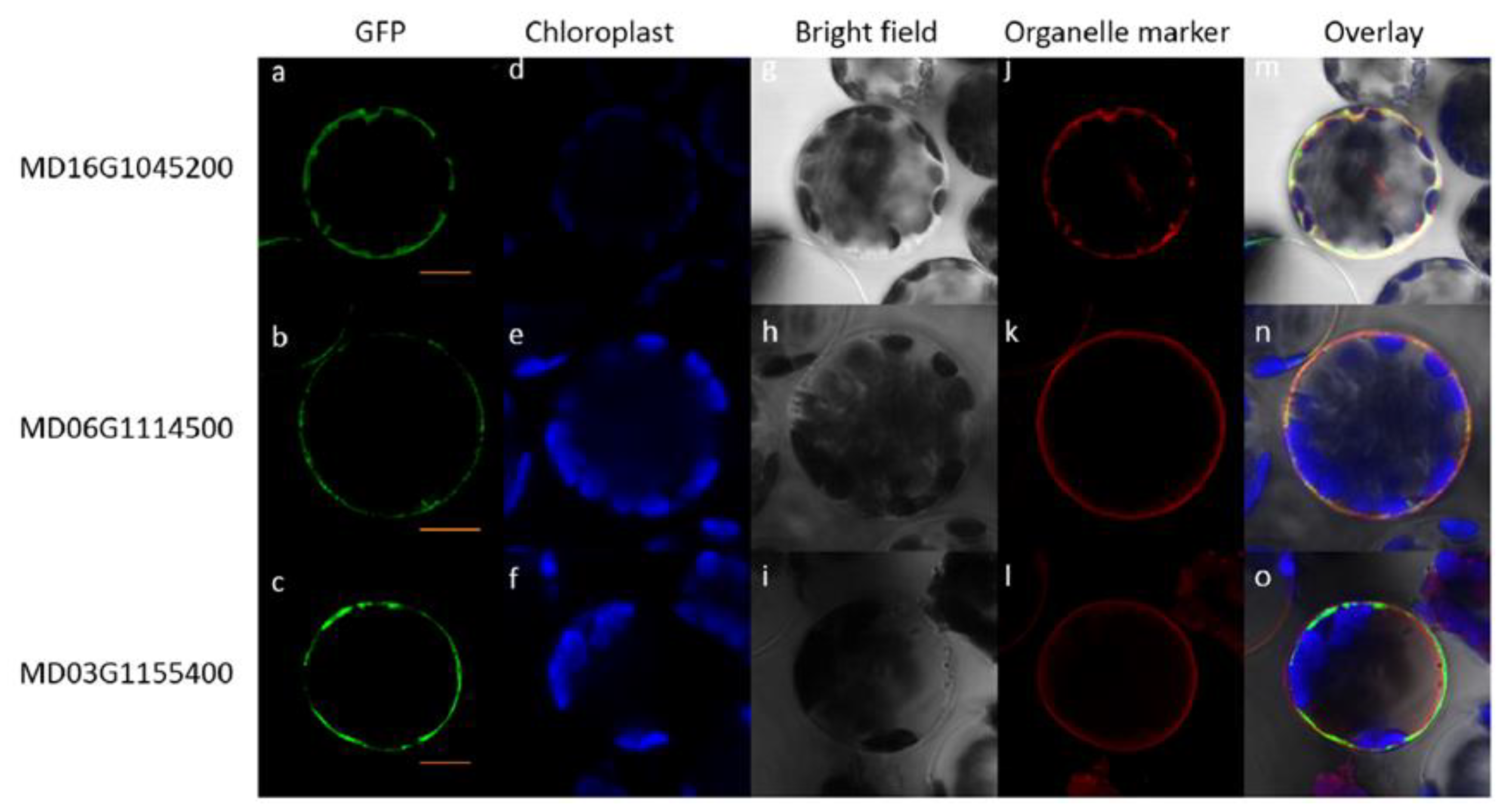
2.6. Different Expression Patterns and Subcellular Positions of ALMT Genes
3. Discussion
4. Materials and Methods
4.1. Identification of ALMT Genes and Phylogenetic Analyses
4.2. Gene Structure and Conserved Motif Analysis of ALMTs
4.3. Syntenic Analysis of ALMTs in Apple
4.4. Gene Duplication and Gene-Loss Estimations
4.5. Detection of Positive Selection
4.6. Detection of Functional Divergence after Gene Duplication
4.7. RNA Isolation and Quantitative RT-PCR (qRT-PCR) Analysis
4.8. Subcellular Localization of Apple ALMTs in Arabidopsis Protoplasts
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Meyer, S.; DeAngeli, A.; Fernie, A.R.; Martinoia, E. Intra-and extra-cellular excretion of carboxylates. Trends Plant Sci. 2010, 15, 40–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, T.; Dreyer, I.; Kochian, L.; Piñeros, M.A. The ALMT family of organic acid transporters in plants and their involvement in detoxification and nutrient security. Front. Plant Sci. 2016, 7, 1488. [Google Scholar] [CrossRef] [PubMed]
- Ryan, P.R.; Skerrett, M.; Findlay, G.P.; Delhaize, E.; Tyerman, S.D. Aluminum activates an anion channel in the apical cells of wheat roots. Proc. Natl. Acad. Sci. USA 1997, 94, 6547–6552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, B.Q.; Liao, L.; Zheng, H.Y.; Chen, J.; Wu, B.H.; Ogutu, C.; Li, S.H.; Korban, S.S.; Han, Y.P. Genes encoding aluminum-activated malate transporter II and their association with fruit acidity in apple. Plant Genome 2015, 8, 1–14. [Google Scholar] [CrossRef]
- Delhaize, E.; Craig, S.; Beaton, C.D.; Bennet, R.J.; Jagadish, V.C.; Randall, P.J. Aluminum tolerance in wheat (Triticumaestivum L.) (I. uptake and distribution of aluminum in root apices). Plant Physiol. 1993, 103, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Delhaize, E.; Ryan, P.R.; Randall, P.J. Aluminum tolerance in wheat (Triticumaestivum L.) (II. aluminum-stimulated excretion of malic acid from root apices). Plant Physiol. 1993, 103, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Zheng, S.J.; Matsumoto, H. Specific secretion of citric acid induced by Al stress in Cassiatora L. Plant Cell Physiol. 1997, 38, 1019–1025. [Google Scholar] [CrossRef]
- Hoekenga, O.A.; Maron, L.G.; Piñeros, M.A.; Cançado, G.M.A.; Shaff, J.; Kobayashi, Y.; Ryan, P.R.; Dong, B.; Delhaize, E.; Sasaki, T.; et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 9738–9743. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Kobayashi, Y.; Sugimoto, M.; Lakshmanan, V.; Iuchi, S.; Kobayashi, M.; Bais, H.P.; Koyama, H. Characterization of the complex regulation of AtALMT1 expression in response to phytohormones and other inducers. Plant Physiol. 2013, 162, 732–740. [Google Scholar] [CrossRef] [PubMed]
- Kovermann, P.; Meyer, S.; Hörtensteiner, S.; Picco, C.; Scholz-Starke, J.; Ravera, S.; Lee, Y.; Martinoia, E. The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J. 2007, 52, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.; Scholz-Starke, J.; DeAngeli, A.; Kovermann, P.; Burla, B.; Gambale, F.; Martinoia, E. Malate transport by the vacuolar AtALMT6 channel in guard cells is subject to multiple regulation. Plant J. 2011, 67, 247–257. [Google Scholar] [CrossRef] [PubMed]
- De Angeli, A.; Zhang, J.; Meyer, S.; Martinoia, E. AtALMT9 is a malate-activated vacuolar chloride channel required for stomatal opening in Arabidopsis. Nat. Commun. 2013, 4, 1804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Angeli, A.; Baetz, U.; Francisco, R.; Zhang, J.; Chaves, M.M.; Regalado, A. The vacuolar channel VvALMT9 mediates malate and tartrate accumulation in berries of Vitis vinifera. Planta 2013, 238, 283–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rensing, S.A. Gene duplication as a driver of plant morphogenetic evolution. Curr. Opin. Plant Biol. 2014, 17, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of gene duplication in plant. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.H.; Han, Z.X.; Jiang, H.Y.; Tian, D.C.; Yang, S.H. Strong positive selection drives rapid diversification of R-genes in Arabidopsis relatives. J. Mol. Evol. 2010, 70, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ding, J.; Zhang, W.; Zhang, Y.L.; Tang, P.; Chen, J.Q.; Tian, D.C.; Yang, S.H. Unique evolutionary pattern of numbers of gramineous NBS-LRR genes. Mol. Genet. Genom. 2010, 283, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhou, H.; Ma, B.Q.; Owiti, A.; Korban, S.S.; Han, Y.P. Divergent evolutionary pattern of sugar transporter genes is associated with the difference in sugar accumulation between grasses and eudicots. Sci. Rep. 2016, 6, 29153. [Google Scholar] [CrossRef] [PubMed]
- Daccord, N.; Celton, J.M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D.; et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhunov, E.D.; Sehgal, S.; Liang, H.; Wang, S.; Akhunova, A.R.; Kaur, G.; Li, W.L.; Forrest, K.L.; See, D.; Simková, H.; et al. Comparative analysis of syntenic genes in grass genomes reveals accelerated rates of gene structure and coding sequence evolution in polyploid wheat. Plant Physiol. 2013, 161, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.S.; Raes, J. Duplication and divergence: The evolution of new genes and old ideas. Annu. Rev. Genet. 2004, 38, 615–643. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Qiao, X.; Zhang, M.; Zhang, S. Genome-wide analysis of aluminum-activated malate transporter family genes in six rosaceae species, and expression analysis and functional characterization on malate accumulation in chinese white pear. Plant Sci. 2018, 274, 451–465. [Google Scholar] [CrossRef]
- Han, Y.P.; Zheng, D.; Vimolmangkang, S.; Khan, M.A.; Beever, J.E.; Korban, S.S. Integration of physical and genetic maps in apple confirms whole-genome and segmental duplications in the apple genome. J. Exp. Bot. 2011, 62, 5117–5130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maere, S.; De Bodt, S.; Raes, J.; Casneuf, T.; Van Montagu, M.; Kuiper, M.; Van de Peer, Y. Modeling gene and genome duplications in eukaryotes. Proc. Nat. Acad. Sci. USA 2005, 102, 5454–5459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Verde, I.; Jenkins, J.; Dondini, L.; Micali, S.; Pagliarani, G.; Vendramin, E.; Paris, R.; Aramini, V.; Gazza, L.; Rossini, L.; et al. The Peach v2.0 release: High-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genom. 2017, 18, 225. [Google Scholar] [CrossRef] [PubMed]
- Chagné, D.; Crowhurst, R.N.; Pindo, M.; Thrimawithana, A.; Deng, C.; Ireland, H.; Fiers, M.; Dzierzon, H.; Cestaro, A.; Fontana, P.; et al. The Draft Genome Sequence of European Pear (Pyrus communis L. ‘Bartlett’). PLoS ONE 2014, 9, e92644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VanBuren, R.; Bryant, D.; Bushakra, J.M.; Vining, K.J.; Edger, P.P.; Rowley, E.R.; Priest, H.D.; Michael, T.P.; Lyons, E.; Filichkin, S.A.; et al. The genome of black raspberry (Rubus occidentalis). Plant J. 2016, 87, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z. PAML 4: A program package for phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteomics Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Gu, X. Maximum likelihood approach for gene family evolution under functional divergence. Mol. Biol. Evol. 2001, 18, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.K.; Cai, X.; Nebenführ, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]





| Gene ID | Animo Acids | MW (kDa) | pI | GRAVY | Instability Index | Aliphatic Index | TMD |
|---|---|---|---|---|---|---|---|
| MD13G1044200 | 521 | 58.60 | 8.30 | 0.066 | 26.07 | 91.71 | 6 |
| MD07G1153600 | 498 | 54.52 | 7.93 | 0.135 | 28.12 | 101.43 | 6 |
| MD13G1044400 | 284 | 31.61 | 8.86 | 0.357 | 25.08 | 104.01 | 6 |
| MD14G1116200 | 546 | 61.23 | 6.69 | −0.125 | 40.61 | 91.43 | 6 |
| MD06G1096000 | 719 | 80.62 | 7.18 | −0.106 | 33.56 | 86.51 | 7 |
| MD14G1135700 | 497 | 54.36 | 7.09 | 0.158 | 27.06 | 96.20 | 7 |
| MD06G1114500 | 488 | 53.70 | 7.88 | 0.148 | 31.09 | 95.16 | 7 |
| MD03G1155400 | 484 | 53.33 | 8.83 | 0.048 | 41.05 | 94.50 | 6 |
| MD11G1173000 | 485 | 53.83 | 9.39 | 0.041 | 36.04 | 95.32 | 6 |
| MD03G1155200 | 812 | 90.91 | 9.55 | −0.174 | 43.50 | 92.60 | 6 |
| MD14G1039500 | 494 | 53.86 | 8.75 | 0.221 | 34.37 | 107.19 | 6 |
| MD12G1040500 | 532 | 57.95 | 8.76 | 0.164 | 32.81 | 104.10 | 6 |
| MD03G1155500 | 467 | 51.96 | 8.39 | 0.046 | 35.46 | 96.30 | 6 |
| MD16G1045200 | 568 | 63.85 | 5.97 | 0.012 | 36.38 | 93.19 | 7 |
| MD16G1045000 | 600 | 66.51 | 6.64 | 0.131 | 28.26 | 94.12 | 7 |
| MD06G1214800 | 603 | 67.64 | 7.92 | −0.095 | 43.33 | 89.40 | 6 |
| MD14G1225700 | 597 | 66.89 | 7.55 | −0.080 | 38.72 | 89.82 | 6 |
| MD06G1032000 | 541 | 60.31 | 8.17 | 0.057 | 34.71 | 98.61 | 6 |
| MD16G1276700 | 522 | 58.68 | 7.99 | −0.007 | 34.82 | 98.30 | 6 |
| MD14G1135900 | 496 | 54.49 | 8.22 | 0.183 | 30.68 | 99.33 | 7 |
| MD03G1266500 | 423 | 46.88 | 8.09 | 0.145 | 39.44 | 106.71 | 6 |
| MD11G1287000 | 433 | 48.22 | 8.31 | 0.164 | 39.89 | 106.03 | 6 |
| MD11G1287100 | 430 | 47.61 | 6.56 | 0.224 | 36.35 | 107.91 | 6 |
| MD00G1017600 | 472 | 51.81 | 8.21 | 0.151 | 43.26 | 97.52 | 6 |
| MD00G1049200 | 485 | 53.69 | 9.28 | 0.066 | 36.40 | 97.94 | 6 |
| Subgroup | Null Hypothesis | Alternative Hypothesis | LRT | ||||
|---|---|---|---|---|---|---|---|
| -In L | ω | -In L | ω1 | ω2 | Statistic | p | |
| I + II | 8024.95 | 0.27 | 8024.38 | 0.25 | 0.29 | 1.14 | >0.05 |
| (I + II) + III | 7860.32 | 0.28 | 7860.08 | 0.28 | 0.30 | 0.48 | >0.05 |
| (I + II + III) + VI | 9497.94 | 0.25 | 9496.27 | 0.26 | 0.21 | 3.34 | >0.05 |
| VI + VII | 3210.45 | 0.22 | 3210.00 | 0.21 | 0.25 | 0.90 | >0.05 |
| (VI + VII) + V | 4121.21 | 0.20 | 4114.04 | 0.22 | 0.08 | 14.34 | <0.01 |
| Subgroup A | Subgroup B | θ-II Value |
|---|---|---|
| I | II | 0.22 ± 0.05 |
| III | −0.28 ± 0.16 | |
| IV | 0.21 ± 0.13 | |
| V | 0.62 ± 0.04 | |
| VI | 0.51 ± 0.05 | |
| VII | 0.87 ± 0.30 | |
| II | III | −0.25 ± 0.11 |
| IV | 0.43 ± 0.04 | |
| V | 0.53 ± 0.07 | |
| VI | 0.65 ± 0.03 | |
| VII | 0.52 ± 0.06 | |
| III | IV | 0.13 ± 0.10 |
| V | 0.46 ± 0.07 | |
| VI | 0.25 ± 0.12 | |
| VII | 0.24 ± 0.11 | |
| IV | V | 0.57 ± 0.03 |
| VI | 0.55 ± 0.04 | |
| VII | 0.21 ± 0.11 | |
| V | VI | 0.52 ± 0.05 |
| VII | 0.45 ± 0.05 | |
| VI | VII | 0.26 ±0.06 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, B.; Yuan, Y.; Gao, M.; Qi, T.; Li, M.; Ma, F. Genome-Wide Identification, Molecular Evolution, and Expression Divergence of Aluminum-Activated Malate Transporters in Apples. Int. J. Mol. Sci. 2018, 19, 2807. https://doi.org/10.3390/ijms19092807
Ma B, Yuan Y, Gao M, Qi T, Li M, Ma F. Genome-Wide Identification, Molecular Evolution, and Expression Divergence of Aluminum-Activated Malate Transporters in Apples. International Journal of Molecular Sciences. 2018; 19(9):2807. https://doi.org/10.3390/ijms19092807
Chicago/Turabian StyleMa, Baiquan, Yangyang Yuan, Meng Gao, Tonghui Qi, Mingjun Li, and Fengwang Ma. 2018. "Genome-Wide Identification, Molecular Evolution, and Expression Divergence of Aluminum-Activated Malate Transporters in Apples" International Journal of Molecular Sciences 19, no. 9: 2807. https://doi.org/10.3390/ijms19092807





