Bacterial Small RNAs in the Genus Herbaspirillum spp.
Abstract
:1. Introduction
2. Results
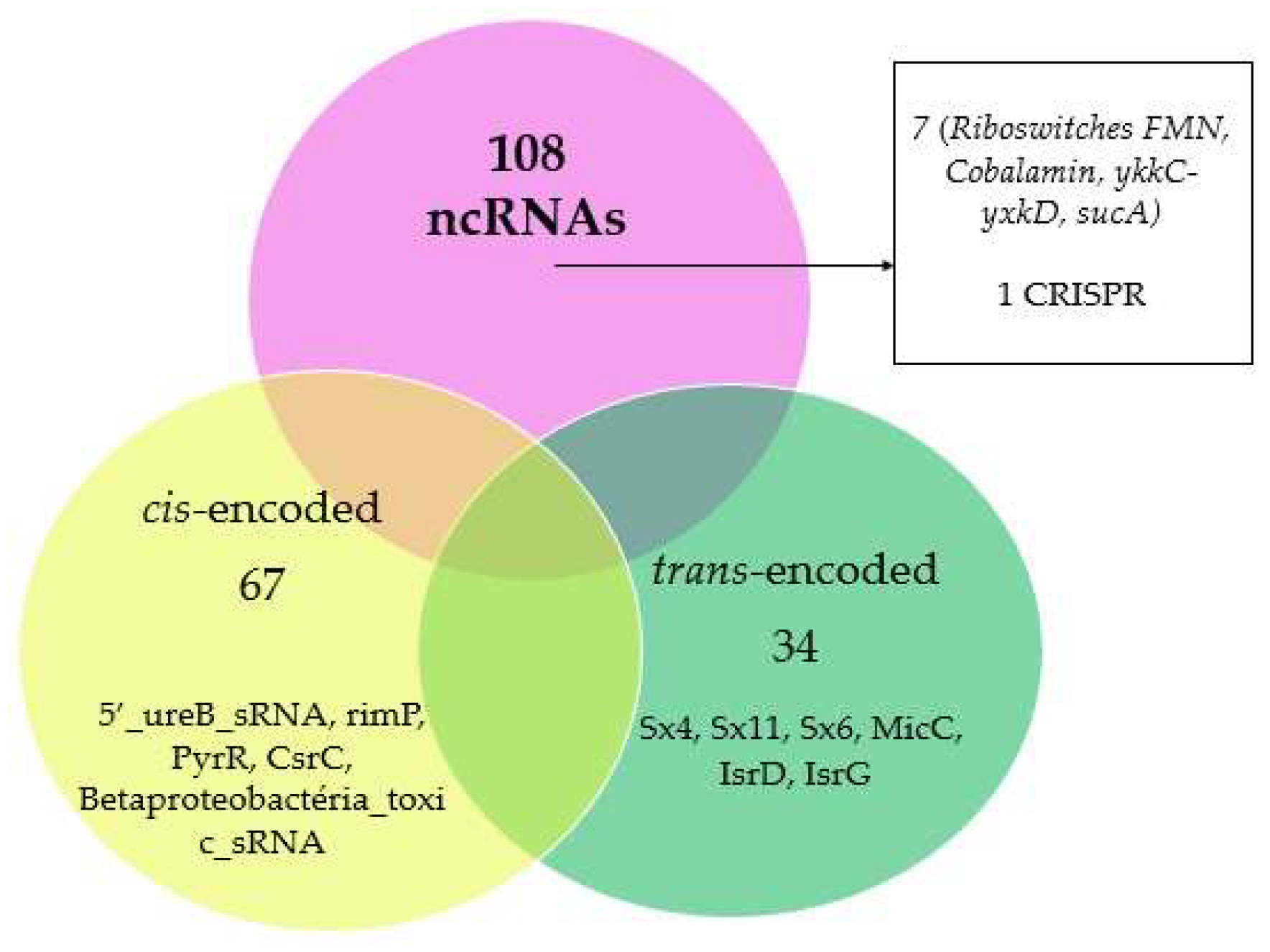
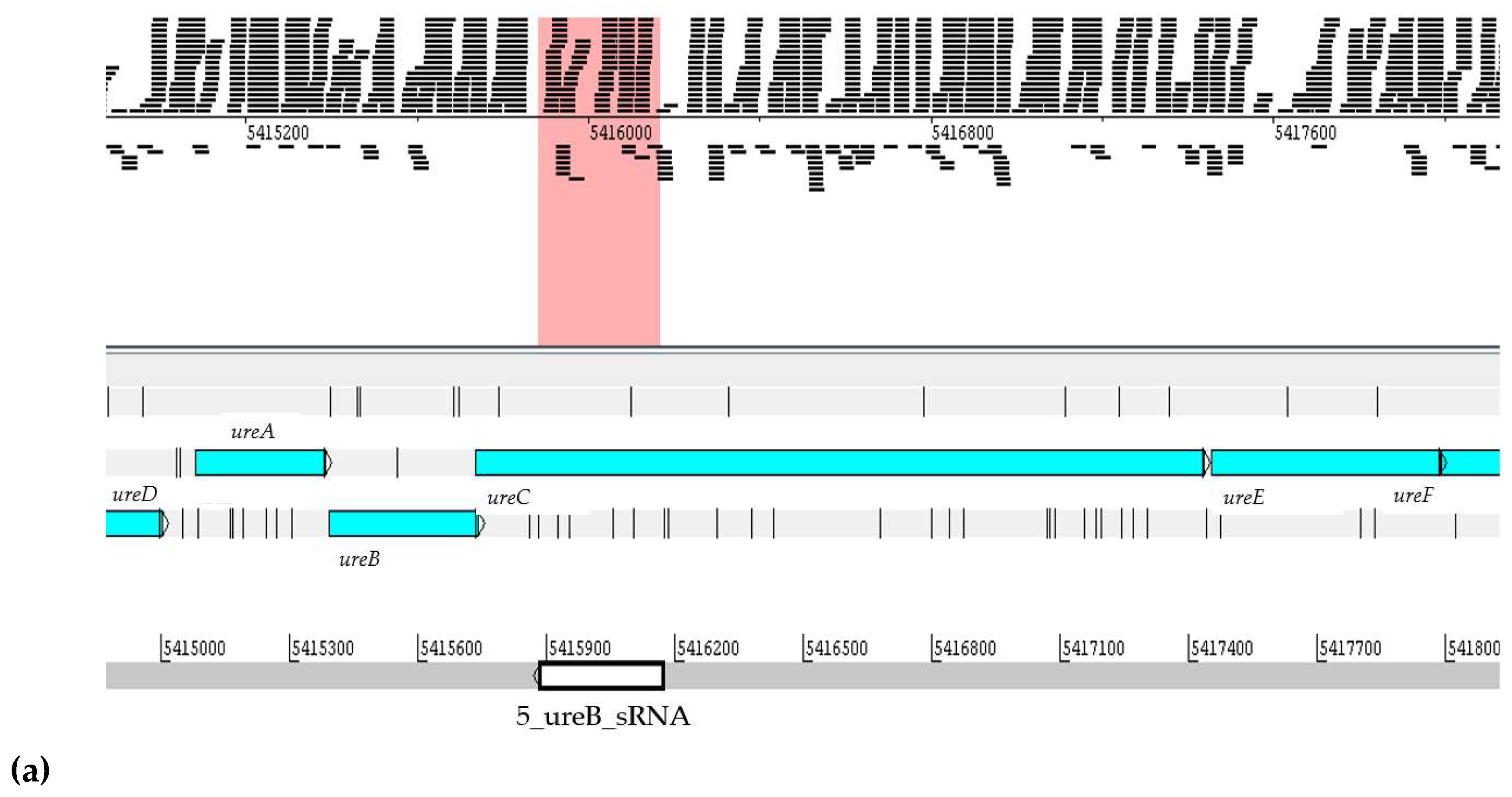
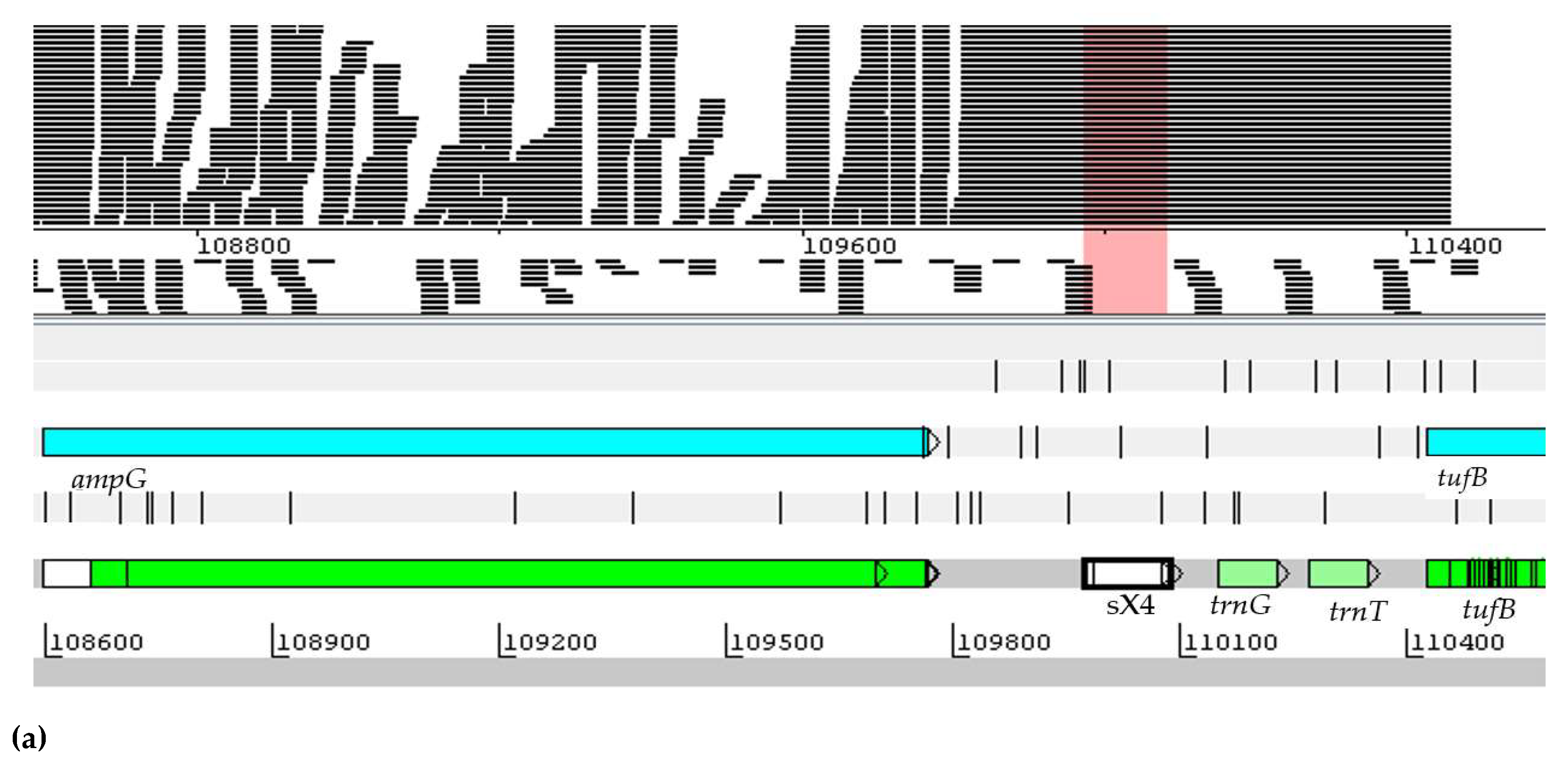
2.1. Prediction of New ncRNAs in the Genus Herbaspirillum spp.
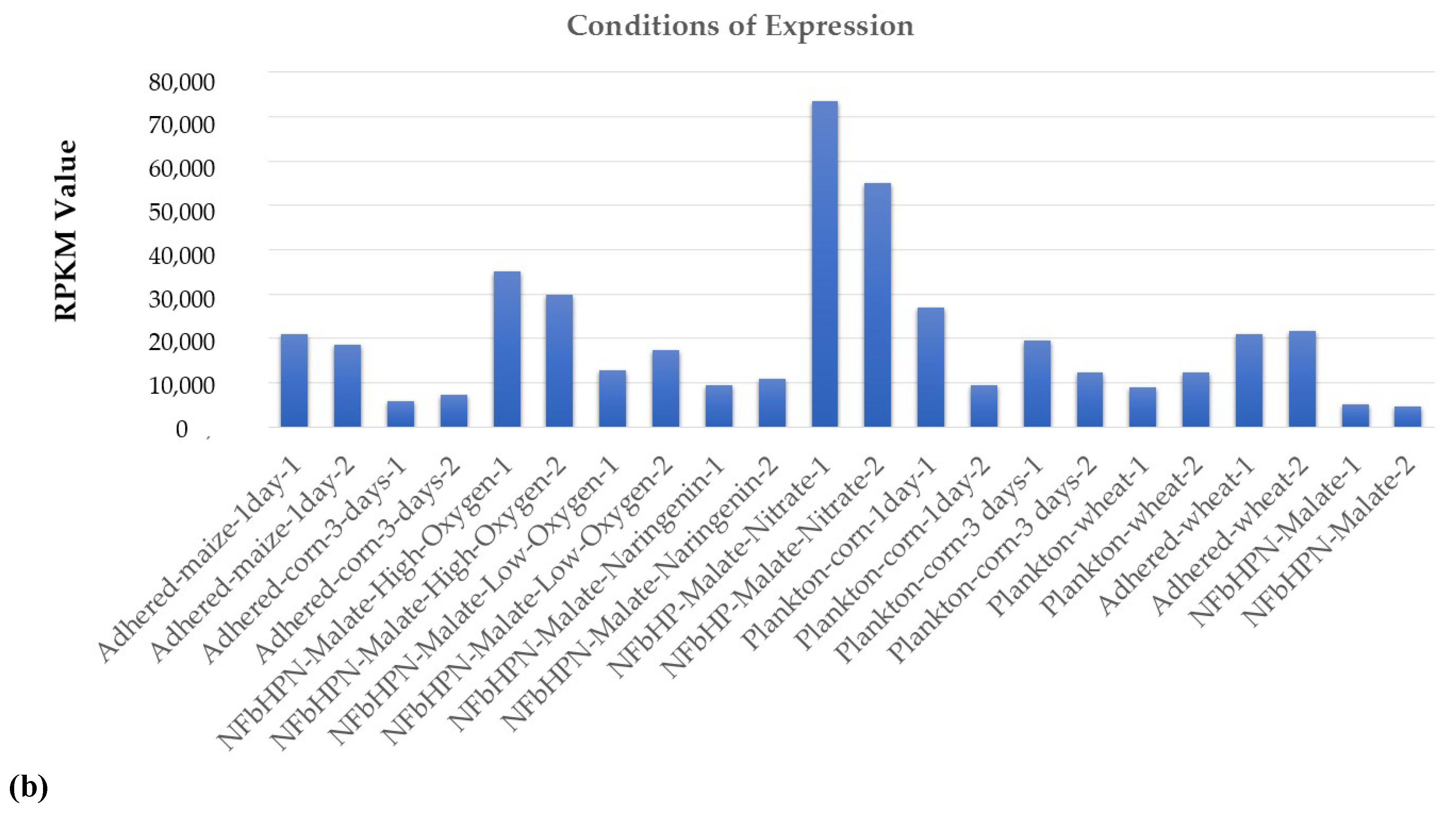
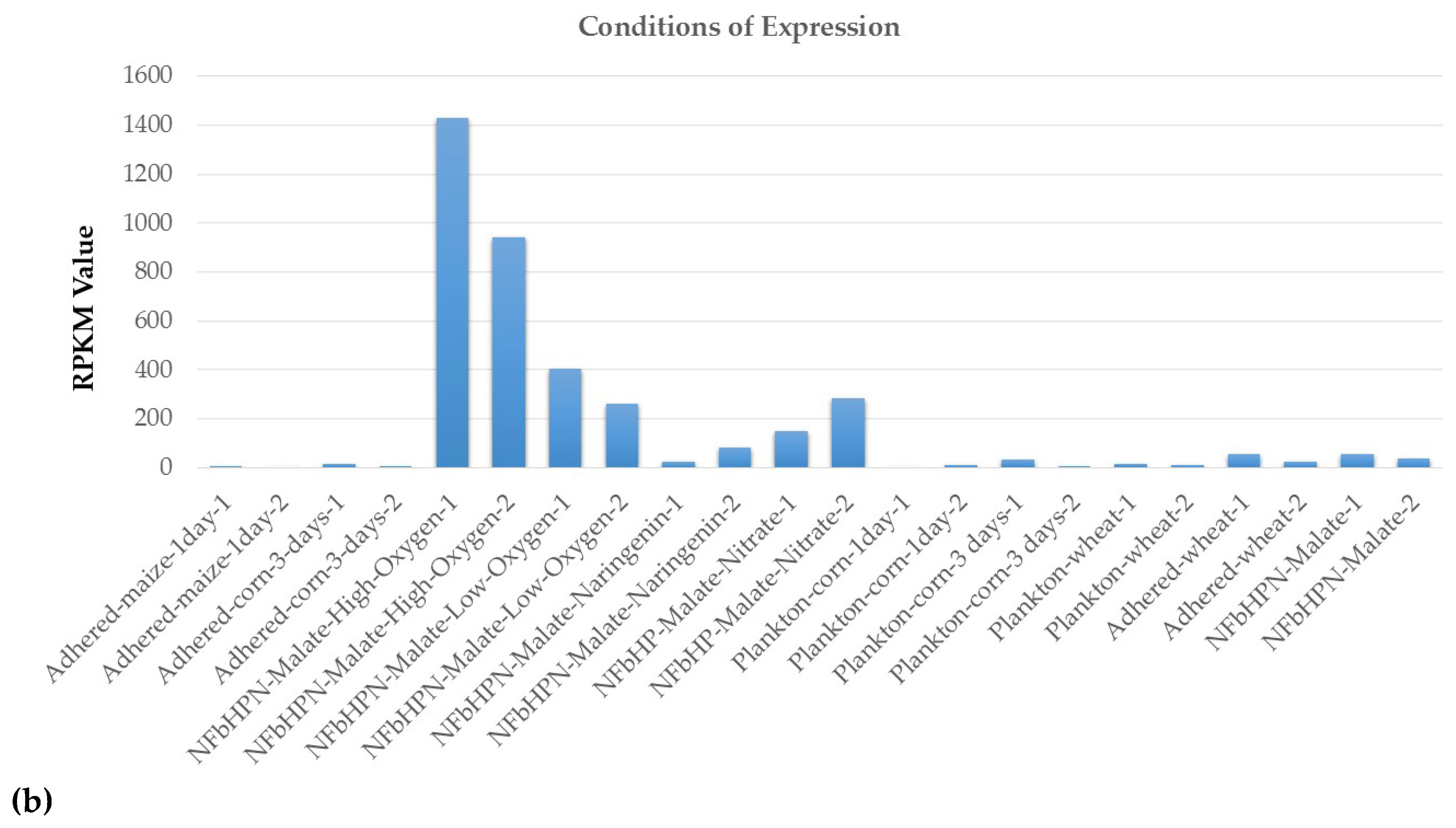
2.2. ncRNAs Differential Expression in RNA-Seq
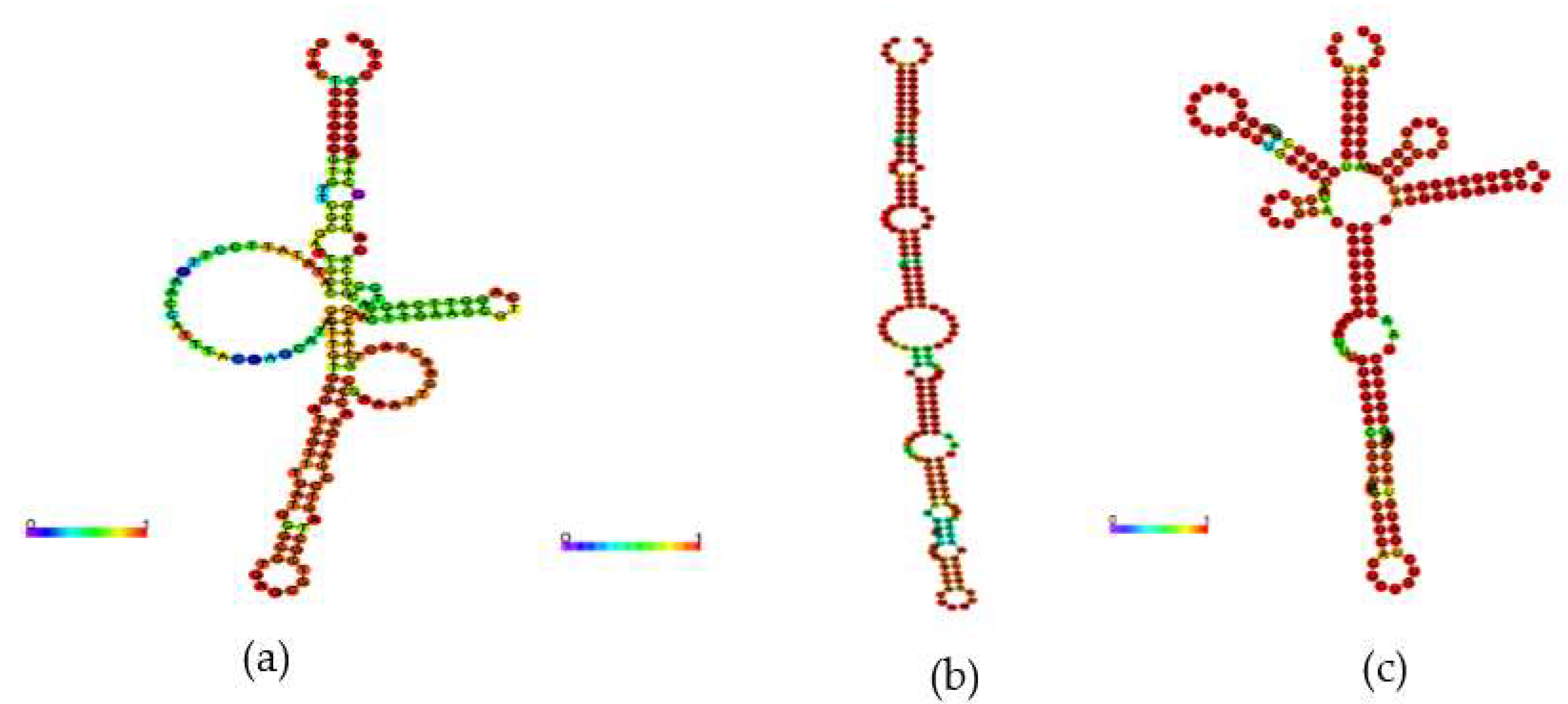
2.3. Comparative Analysis of the Secondary Structure of 6S of H. seropedicae SmR1
2.4. Candidate sRNA Target Identification
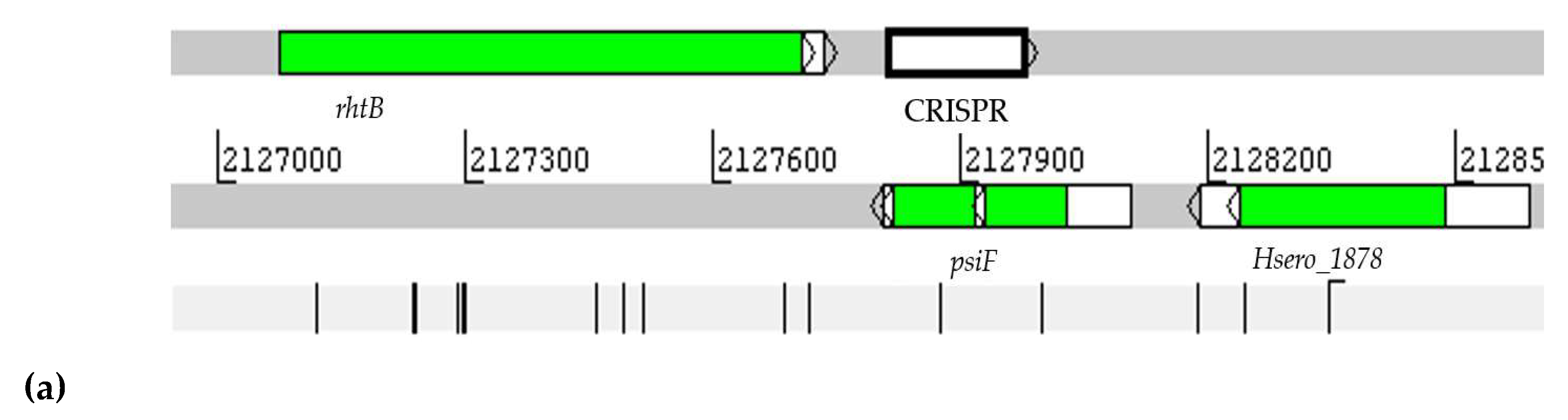
2.5. Prediction of CRISPR
3. Discussion
4. Materials and Methods
4.1. Genomes of Bacteria of the Genus Herbaspirillum
4.2. RNA-Seq Data
4.3. Prediction of ncRNAs in Silico
4.4. ncRNA Sequence Annotation
4.5. Mapping and Visualization of Sequence Reads
4.6. Transcriptome Analysis
4.7. Target Prediction
4.8. Identifying CRISPR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Baldani, J.I.; Baldani, V.L.D.; Seldin, L.; Döbereiner, J. Characterization of Herbaspirillum seropedicae nov. sp. a root associated nitrogen fixing bacterium. Int. J. Syst. Evol. Microbiol. 1986, 36, 86–93. [Google Scholar] [CrossRef]
- Baldani, J.I.; Pot, B.; Kirchhof, G.; Falsen, E.; Baldani, V.L.D.; Olivares, F.L.; Hoste, B.; Kersters, K.; Hartmann, A.G.; Dobereiner, J. Emended description of Herbaspirillum; inclusion of Pseudomonas rubrisubalbicans, a mild plant pathogen, as Herbaspirillum rubrisubalbicans comb nov; and classification of a group of clinical isolates (EF group 1) as Herbaspirillum species 3. Int. J. Syst. Evol. Microbiol. 1996, 46, 802–810. [Google Scholar]
- Ding, L.; Yakota, A. Proposals of Curvibacter gracilis gen. nov., sp. nov. and Herbaspirillum putei sp. nov. for bacterial strains isolated from well water and reclassification of [Pseudomonas] huttiensis, [Pseudomonas] lanceolata, [Aquaspirillum] delicatum and [Aquaspirillum] autotrophicum as Herbaspirillum huttiense comb. nov., Curvibacter lanceolatus comb. nov., Curvibacter delicatus comb. nov. and Herbaspirillum autotrophicum comb. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 2223–2230. [Google Scholar] [PubMed]
- Kirchhof, G.; Eckert, B.; Stoffels, M.; Baldani, J.I.; Reis, V.M.; Hartmann, A. Herbaspirillum frisingense sp. Nov., a new nitrogen-fixing bacterial species that occurs in c4-fibre plants. Int. J. Syst. Evol. Microbiol. 2001, 51, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Valverde, V.B.A.; Velázquez, E.; Gutiérrez, C.; Cervantes, E.; Ventosa, A.; Igual, J.M. Herbaspirillum lusitanum sp. nov., a novel nitrogen-fixing bacterium associated with root nodules of Phaseolus vulgaris. Int. J. Syst. Evol. Microbiol. 2003, 53, 1979–1983. [Google Scholar] [PubMed]
- Im, W.T.; Bae, H.S.; Yokota, A.; Lee, S.T. Herbaspirillum chlorophenolicum sp. nov., a 4-chlorophenol-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 851–855. [Google Scholar] [CrossRef] [PubMed]
- Rothballer, M.; Schmid, M.; Klein, I.; Gattinger, A.; Grundmann, S.; Hartmann, A. Herbaspirillum hiltneri sp. nov., isolated from surface-sterilized wheat roots. Int. J. Syst. Evol. Microbiol. 2006, 56, 1341–1348. [Google Scholar]
- Jung, S.Y.; Lee, M.H.; Oh, T.K.; Yoon, J.H. Herbaspirillum rhizosphaerae sp. nov., isolated from rhizosphere soil of Allium victorialis var. platyphyllum. Int. J. Syst. Evol. Microbiol. 2007, 57, 2284–2288. [Google Scholar] [CrossRef] [Green Version]
- Dobritsa, A.P.; Reddy, M.C.; Samadpour, M. Reclassification of Herbaspirillum putei as a later heterotypic synonym of Herbaspirillum huttiense, with the description of H. huttiense subsp. huttiense subsp. nov. and H. huttiense subsp. putei subsp. nov., comb. nov., and description of Herbaspirillum aquaticum sp. nov. Int. J. Syst. Evol. Microbiol. 2009, 60, 1418–1426. [Google Scholar]
- Lagier, J.C.; Gimenez, G.; Robert, C.; Raoult, D.; Fournier, P.E. Non-contiguous finished genome sequence and description of Herbaspirillum massiliense sp. Stand. Genom. Sci. 2012, 7, 200–209. [Google Scholar] [CrossRef]
- Carro, L.; Rivas, R.; León-Barrios, M.; González-Tirante, M.; Velázquez, M.; Valverde, A. Herbaspirillum canariense sp. nov., Herbaspirillum aurantiacum sp. nov., and Herbaspirillum soli sp. nov., isolated from volcanic mountain soil, and emended description of the genus. Int. J. Syst. Evol. Microbiol. 2012, 62, 1300–1306. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.C.; Paludo, K.S.; Dallagassa, C.B.; Surek, M.; Pedrosa, F.O.; Souza, E.M.; Cruz, L.M.; Lipuma, J.J.; Zanata, S.M.; Rego, F.G.; et al. Biochemical characteristics, adhesion, and cytotoxicity of environmental and clinical isolates of Herbaspirillum. J. Clin. Microbiol. 2015, 53, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Baldani, J.I.; Baldani, V.L.D.; Sampaio, M.J.A.M.; Döbereiner, J. A fourth azospirillum species from cereal roots. In Anais da Academia Brasileira de Ciência; Brazilian Academy of Sciences: Rio de Janeiro, Brasil, 1984; Volume 56, pp. 351–365. [Google Scholar]
- James, E.K.; Olivares, F.L.; Baldani, J.I.; Dobereiner, J. Herbaspirillum, an endophytic diazotroph colonizing vascular tissue in leaves of Sorghum Bicolor L. Moench. J. Exper. Bot. 1997, 48, 785–797. [Google Scholar] [CrossRef]
- Baldani, V.L.D.; Baldani, J.I.; Olivares, F.L.; Döbereiner, J. Identification and ecology of Herbaspirillum seropedicae and the closely related Pseudomoas rubusubalbicas. Symbiosis 1992, 13, 65–73. [Google Scholar]
- Döbereiner, J.; Baldani, V.L.D.; Baldani, J.I. Como isolar e identificar bactérias diazotróficas de plantas não-leguminosas; EMBRAPA-SPI: Brasília, Brazil, 1995; p. 60. [Google Scholar]
- Olivares, F.L.; Baldani, V.L.D.; Reis, V.M.; Baldani, J.I.; Döbereiner, J. Occurrence of the endophytic diazotrophs Herbaspirillum spp. in roots, stems, and leaves, predominantly of Gramineae. Biol. Fertil. Soils. 1996, 21, 197–200. [Google Scholar] [CrossRef]
- Storz, G.; Altuvia, S.; Wassarman, K.M. An abundance of RNA regulators. Annu. Rev. Biochem. 2005, 74, 199–217. [Google Scholar] [CrossRef]
- Liu, J.M.; Camilli, A. A broadening world of bacterial small RNAs. Curr. Opin. Microbiol. 2010, 13, 18–23. [Google Scholar] [CrossRef] [Green Version]
- Andersen, J.; Delihas, N.; Ikenaka, K.; Green, P.J.; Pines, O.; Ilercil, O.; Inouye, M. The isolation and characterization of RNA coded by the micF gene in Escherichia coli. Nucleic Acids Res. 1987, 15, 2089–2101. [Google Scholar] [CrossRef]
- Ermolaeva, M.D.; Khalak, H.G.; White, O.; Smith, H.O.; Salzberg, S.L. Prediction of Transcription Terminators in Bacterial Genomes. Mol. Biol. 2000, 301, 27–33. [Google Scholar] [CrossRef]
- Argaman, L.; Hershberg, R.; Vogel, J.; Bejerano, G.; Wagner, E.G.H.; Margalit, H.; Altuvia, S. Novel small RNA-encoding genes in the intergenic region of Escherichia coli. Current Biology 2001, 11, 941–950. [Google Scholar] [CrossRef]
- Ciampi, M.S. Rho-dependent terminators and transcription termination. Microbiology 2006, 152, 2515–2528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wassarman, K.M.; Kiley, P.J. Global approaches for finding small RNA and small open reading frame functions. J. Bacteriol. 2010, 192, 26–28. [Google Scholar] [CrossRef] [PubMed]
- Arnvig, K.B.; Cortes, T.; Young, D.B. Noncoding RNA in Mycobacteria. Microbiol. Spectrum 2014, 2, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Updegrove, T.B.; Zhang, A.; Storz, G. Hfq: The flexible RNA matchmaker. Curr. Opin. Microbiol. 2016, 30, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, C.L.C.; Narra, H.P.; Rojas, M.; Sahni, A.; Patel, J.; Khanipov, K.; Wood, T.G.; Fofanov, Y.; Sahni, S.K. Bacterial small RNAs in the Genus Rickettsia. BMC Genomics 2015, 16, 1075. [Google Scholar]
- Pichon, C.; Felden, B. Proteins that interact with bacterial small RNA regulators. Fems. Microbiol. Rev. 2007, 31, 614–625. [Google Scholar] [CrossRef] [Green Version]
- Livny, J.; Waldor, M.K. Identification of small RNAs in diverse bacterial species. Curr. Opin. Microbiol. 2007, 10, 1096–1101. [Google Scholar] [CrossRef]
- Hüttenhofer, A.; Schattner, P.; Polacek, P. Non-coding RNAs: Hope or hype? Trends Genet. 2005, 21, 289–297. [Google Scholar] [CrossRef]
- Brantl, S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007, 10, 102–109. [Google Scholar] [CrossRef]
- Sittka, A.; Lucchini, S.; Papenfort, K.; Sharma, C.M.; Rolle, K.; Binnewies, T.T.; Hinton, J.C.; Vogel, J. Deep sequencing analysis of small non coding RNA and mRNA targets of the global post-transcriptional regulator Hfq. PLoS Genet. 2008, 224, 1–20. [Google Scholar]
- Lorenz, R.; Bernhart, S.H.; Höner, Z.; Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. Vienna RNA Package 2.0. Algorithms. Mol. Biol. 2011, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Rossi, C.C.; Bossé, J.T.; Li, Y.; Witney, A.A.; Gould, K.A.; Langford, P.R.; Bazzolli, D.M. A computational strategy for the search of regulatory small RNAs in Actinobacillus pleuropneumoniae. RNA 2016, 22, 1373–1385. [Google Scholar] [CrossRef] [PubMed]
- Repoila, F.; Darfeuille, F. Small regulatory non-coding RNAs in bacteria: Physiology and mechanistic aspects. Biol. Cell 2009, 101, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Faner, M.A.; Feig, A.L. Identifying and characterizing Hfq–RNA interactions. Methods 2013, 63, 144–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by small RNAs in bacteria: Expanding frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Waters, L.S.; Storz, G. Regulatory RNAs in Bacteria. Cell 2009, 136, 615–628. [Google Scholar] [CrossRef] [Green Version]
- Delihas, N. Regulation of gene expression by trans-encoded antisense RNAs. Mol. Microbiol. 1995, 15, 411–414. [Google Scholar] [CrossRef]
- Winkler, W.C. Riboswitches and the role of noncoding RNAs in bacterial metabolic control. Curr. Opin. Chem. Biol. 2005, 9, 594–602. [Google Scholar] [CrossRef]
- Meyer, M.M.; Hammond, M.C.; Salinas, Y.; Roth, A.; Sudarsan, N.; Breaker, R.R. Challenges of ligand identification for riboswitch candidates. RNA Biol. 2011, 8, 5–10. [Google Scholar] [CrossRef] [Green Version]
- Izar, B.; Mraheil, M.A.; Hain, T. Identification and Role of Regulatory Non-Coding RNAs in Listeria monocytogenes. Int. J. Mol. Sci. 2011, 12, 5070–5079. [Google Scholar] [CrossRef]
- Jiang, W.; Fang, L.; Inouye, M. The role of the 5′-end untranslated region of the mRNA for CspA, the major cold-shock protein of Escherichia coli, in cold-shock adaptation. J. Bacteriol. 1996, 178, 4919–4925. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K.; Inouye, M. Growth-phase-dependent expression of cspD, encoding a member of the CspA family in Escherichia coli. J. Bacteriol. 1997, 179, 5126–5130. [Google Scholar] [CrossRef] [PubMed]
- Bonato, P. Genes do metabolismo de Nitrato em Herbaspirillum seropedicae: Regulação transcricional e análise funcional. Dissertação (Mestrado em Bioquímica)—Setor de Ciências Biológicas. Universidade Federal do Paraná: Curitiba, Brazil, 2012. [Google Scholar]
- SCHMIDTKE, C.; Findeiss, S.; Sharma, C.M.; Kuhfuss, J.; Hoffmann, S.; Vogel, J.; Stadler, P.F.; Bonas, U. Genome-wide transcriptome analysis of the plant pathogen Xanthomonas identifies sRNAs with putative virulence functions. Nucleic Acids Res. 2012, 40, 2020–2031. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, A.; Blyn, L.B.; Storz, G. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 2004, 186, 6689–6697. [Google Scholar] [CrossRef] [PubMed]
- Wasserman, K.M. 6S RNA, a Global Regulator of Transcription. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Higgins, D.G.; Sharp, P.M. CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene 1988, 73, 237–244. [Google Scholar] [CrossRef]
- Nakamura, K.; Miyamoto, H.; Suzuma, S.; Sakamoto, T.; Kawai, G.; Yamane, K. Minimal functional structure of Escherichia coli 4.5 S RNA required for binding to elongation factor G. J. Biol. Chem. 2001, 276, 22844–22849. [Google Scholar] [CrossRef] [PubMed]
- Jagath, J.R.; Matassova, N.B.; Leeuw, D.E.; Warnecke, J.M.; Lentzen, G.; Rodnina, M.V.; Luirink, J.; Wintermeyer, W. Important role of the tetraloop region of 4.5S RNA in SRP binding to its receptor FtsY. RNA 2001, 2, 293–301. [Google Scholar] [CrossRef]
- Gu, S.Q.; Jöckel, J.; Beinker, P.; Warnecke, J.; Semenkov, Y.P.; Rodnina, M.V.; Wintermeyer, W. Conformation of 4.5S RNA in the signal recognition particle and on the 30S ribosomal subunit. RNA 2005, 11, 1374–1384. [Google Scholar] [CrossRef] [Green Version]
- Buskiewicz, I.; Kubarenko, A.; Peske, F.; Rodnina, M.V.; Wintermeyer, W. Domain rearrangement of SRP protein Ffh upon binding 4.5S RNA and the SRP receptor FtsY. RNA 2005, 11, 947–957. [Google Scholar] [CrossRef] [Green Version]
- MOJICA, F.J.; Díez-Villaseñor, C.; Soria, E.; Juez, G. Biological significance of a family of regularly spaced repeats in the genomes of archaea, bacteria and mitochondria. Mol. Microbiol. 2000, 36, 244–246. [Google Scholar] [CrossRef] [PubMed]
- Jansen, G.; Leberer, E.; Thomas, D.Y.; Whiteway, M. Use of dominant negative mutations in analysis of G protein function in Saccharomyces cerevisiae. Methods Enzymol. 2002, 344, 82–91. [Google Scholar] [PubMed]
- Barrangou, R.; Marraffini, L.A. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol. Cell 2014, 54, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Hale, C.R.; Majumdar, S.; Elmore, J.; Pfister, N.; Compton, M.; Olson, S.; Resch, A.M.; Glover, C.V.R.D.; Graveley, B.R.; Terns, R.M.; et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell 2012, 45, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Estrella, M.A.; Kuo, F.T.; Bailey, S. RNA-activated DNA cleavage by the Type III-B CRISPR-Cas effector complex. Genes. Dev. 2016, 30, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.W.; Zhu, Y.; Staals, R.H.; Kornfeld, J.E.; Shinkai, A.; van der Oost, J.; Nogales, E.; Doudna, J.A. Structural biology. Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science 2015, 348, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, X.; Ma, J.; Li, Z.; You, L.; Wang, J.; Wang, M.; Zhang, X.; Wang, Y. The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a. Cell 2017, 170, 714–726. [Google Scholar] [CrossRef] [PubMed]
- Carter, E.L.; Boer, J.L.; Farrugia, M.A.; Flugga, N.; Towns, C.L.; Hausinger, R.P. Function of UreB in Klebsiella aerogenes urease. Biochemistry 2011, 50, 9296–9308. [Google Scholar] [CrossRef]
- Wen, Y.; Feng, J.; Sachs, G. Helicobacter pylori 5 ureB-sRNA, a cis-encoded antisense small RNA, negatively regulates ureAB. J. Bacteriol. 2013, 195, 444–452. [Google Scholar] [CrossRef]
- Abendroth, U.; Schmidtke, C.; Bonas, U. Small non-coding RNAs in plant-pathogenic Xanthomonas. RNA Biol. 2014, 11, 457–463. [Google Scholar] [CrossRef]
- Dam, S.; Pagès, J.M.; Masi, M. Dual regulation of the small RNA MicC and the quiescent porin Omp in response to antibiotic stress in Escherichia coli. Antibiotics 2017, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Ortega, A.D.; Gonzalo-Asensio, J.; García-Del Portillo, F. Dynamics of Salmonella small RNA expression in non-growing bacteria located inside eukaryotic cells. RNA Biol. 2012, 4, 469–488. [Google Scholar] [CrossRef] [PubMed]
- Padalon-Brauch, G.; Hershberg, R.; Elgrably-Weiss, M.; Baruch, K.; Rosenshine, I.; Margalit, H.; Altuvia, S. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res. 2008, 36, 1913–1927. [Google Scholar] [CrossRef] [PubMed]
- Kery, M.B.; Feldman, M.; Livny, J.; Tjaden, B. TargetRNA2: Identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res. 2014, 42, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Zheng, P.; Delihas, N. Secondary structures of Escherichia coli antisense MicF RNA, the 5-end of the target OmpF mRNA, and the RNA/RNA duplex. Biochemistry 1995, 34, 3621–3631. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, E.P.; Kolbe, D.L.; Eddy, S.R. Infernal 1.0: Inference of RNA alignments. Bioinformatics 2009, 25, 1335–1337. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [Green Version]
- Abreu-Goodger, C.; Merino, E. RibEx: A web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res. 2005, 1, 690–692. [Google Scholar] [CrossRef]
- Raghavan, R.; Groisman, E.A.; Ochman, H. Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res. 2011, 21, 1487–1497. [Google Scholar] [CrossRef]
- David, M.; Dzamba, M.; Lister, D.; Ilie, L.; Brudno, M. SHRiMP2: Sensitive yet practical SHort read mapping. Bioinformatics 2011, 27, 1011–1012. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Böhme, U.; Otto, T.D.; Parkhill, J.; Berriman, M. BamView: Viewing mapped read alignment data in the context of the reference sequence. Bioinformatics 2010, 26, 676–677. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; Mccue, K.; Schaeffer, L.; Wold, B. Mapping and Quantifying Mammalian Transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Tjaden, B.; Goodwin, S.S.; Opdyke, J.A.; Guillier, M.; Fu, D.X.; Gottesman, S.; Storz, G. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006, 34, 2791–2802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tjaden, B. TargetRNA: A tool for predicting targets of small RNA action in bacteria. Nucleic Acids Res. 2008, 36, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Backofen, R.; Hess, W.R. Computational prediction of sRNAs and their targets in bacteria. RNA Biol. 2010, 7, 1–10. [Google Scholar] [CrossRef]
- Lange, S.J.; Alkhnbashi, O.S.; Rose, D.; Will, S.; Backofen, R. CRISPRmap: An automated classification of repeat conservation in prokaryotic adaptive immune systems. Nucleic Acids Res. 2013, 41, 8034–8044. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nuclei. Acids Res. 2007, 35 (Suppl. 2), W52–W57. [Google Scholar] [CrossRef]



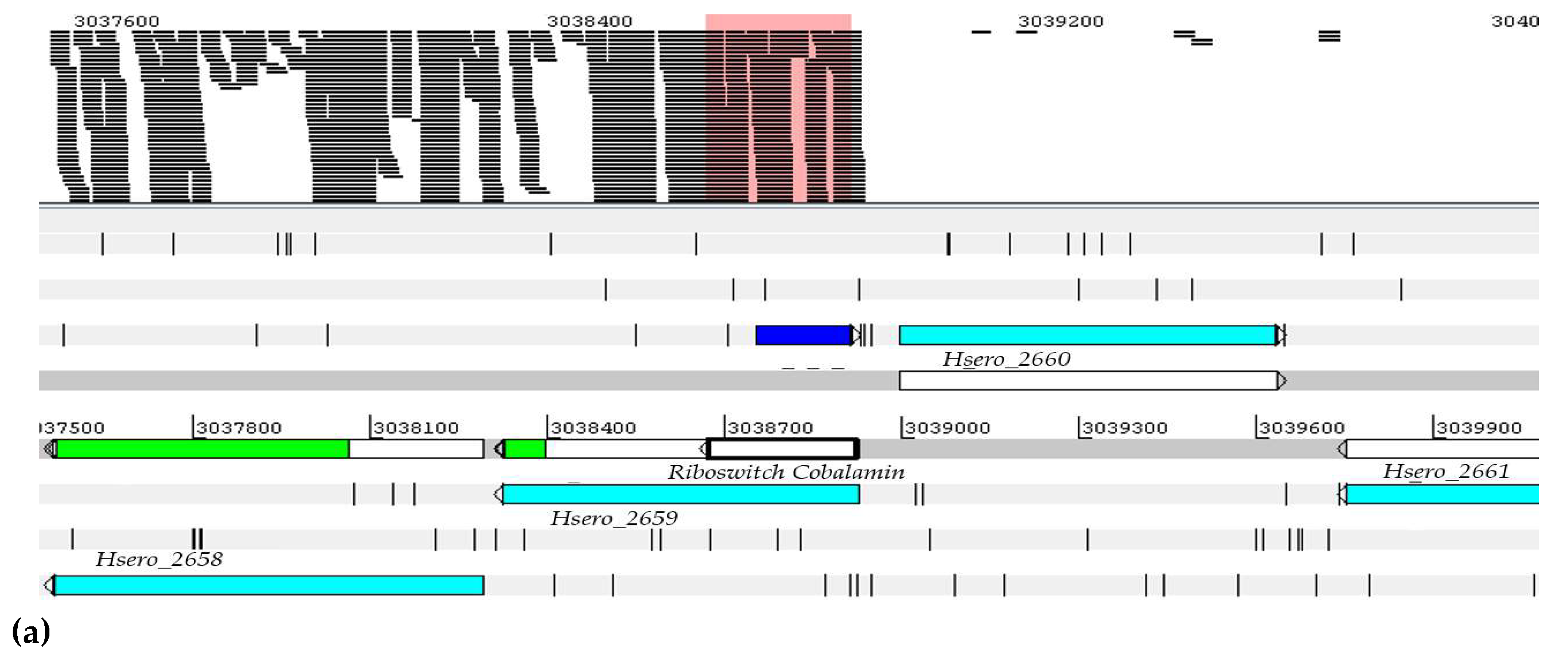
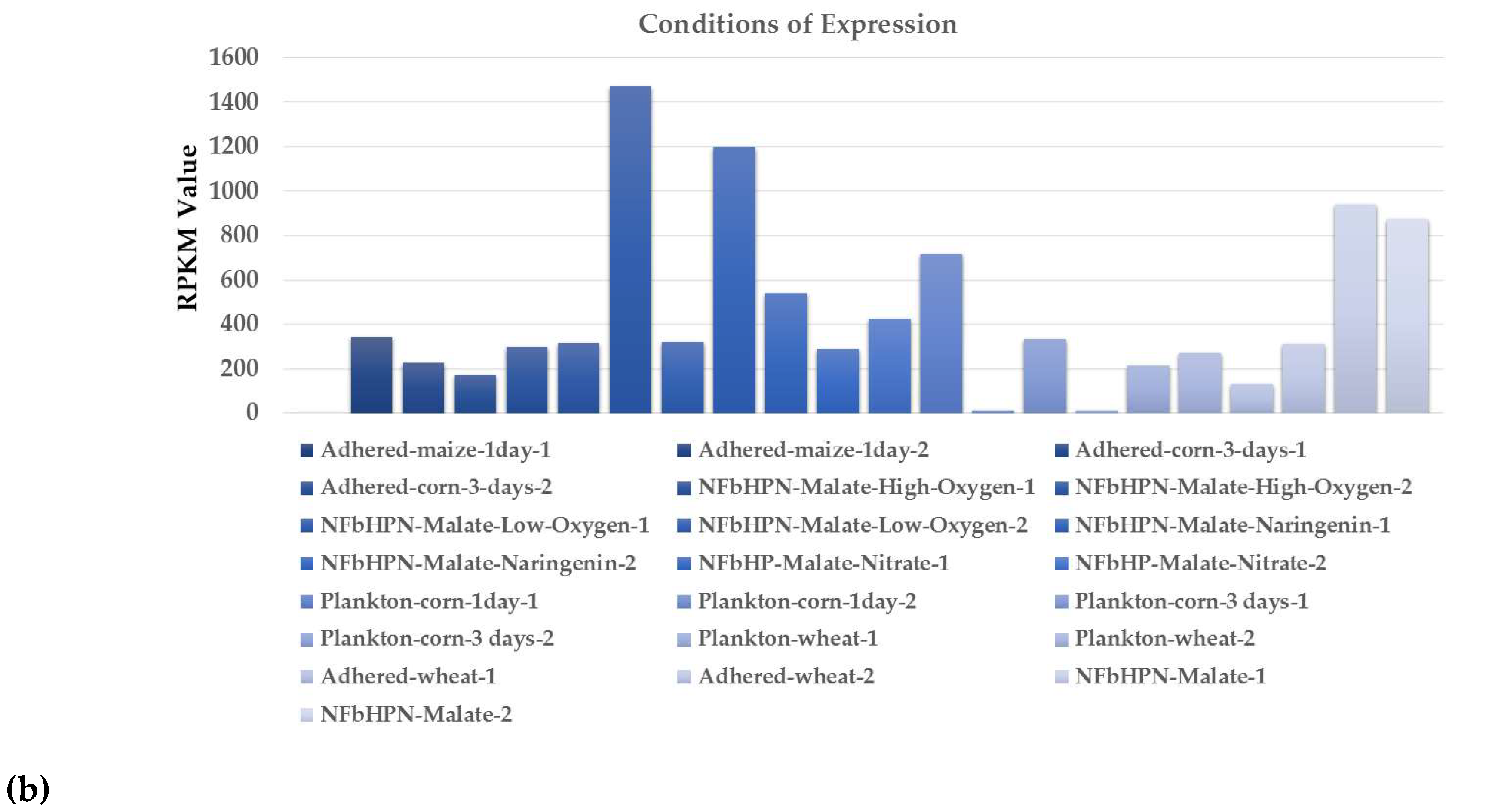
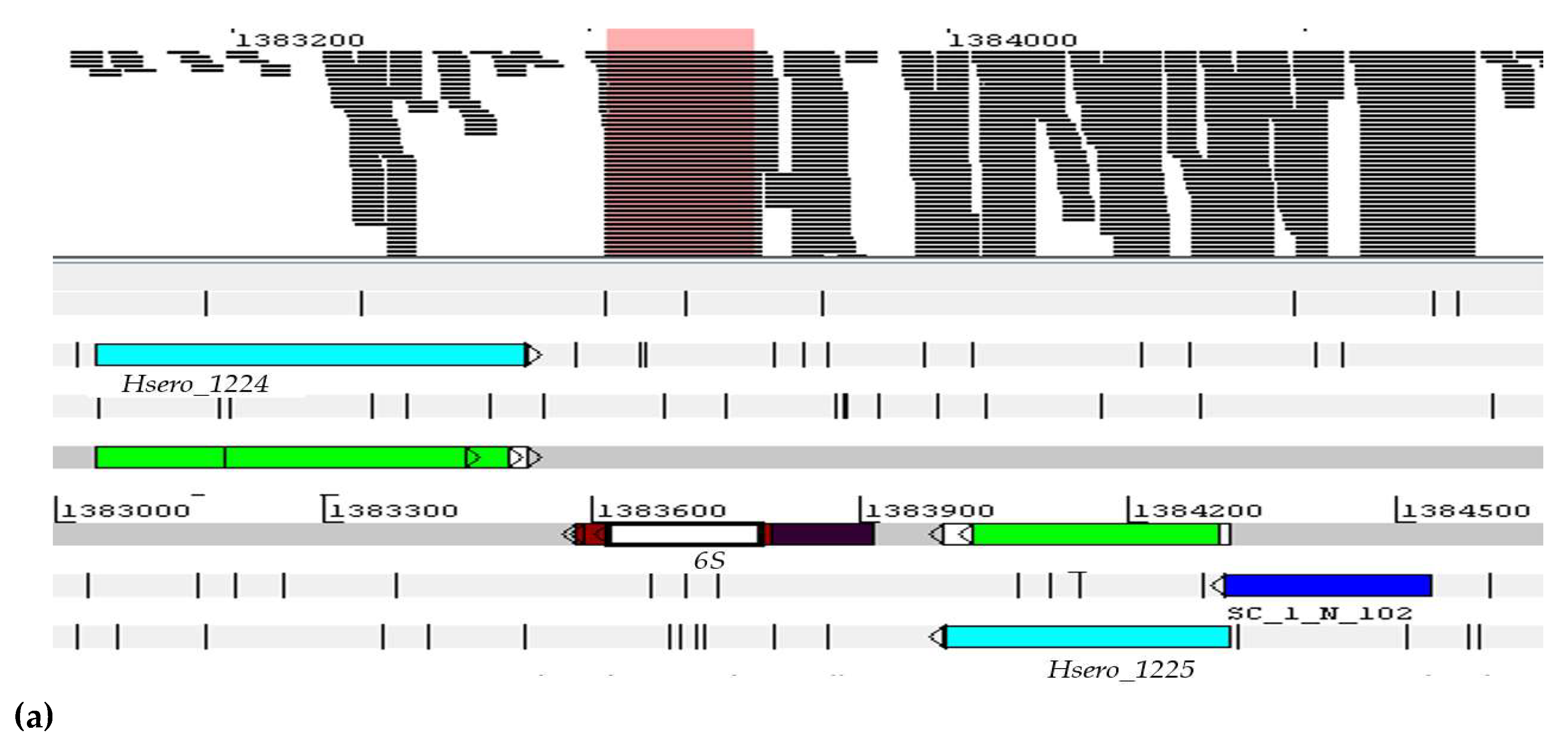
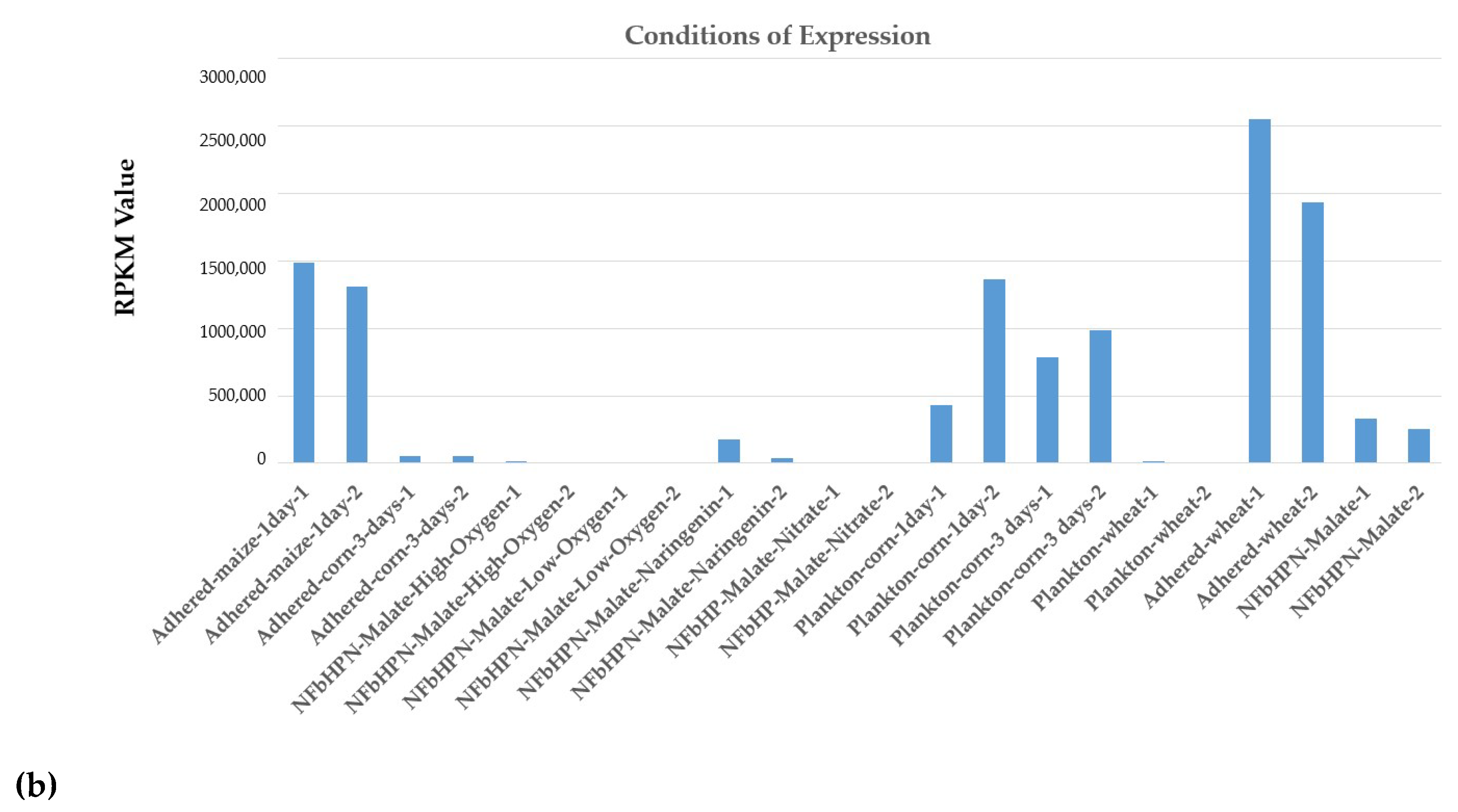



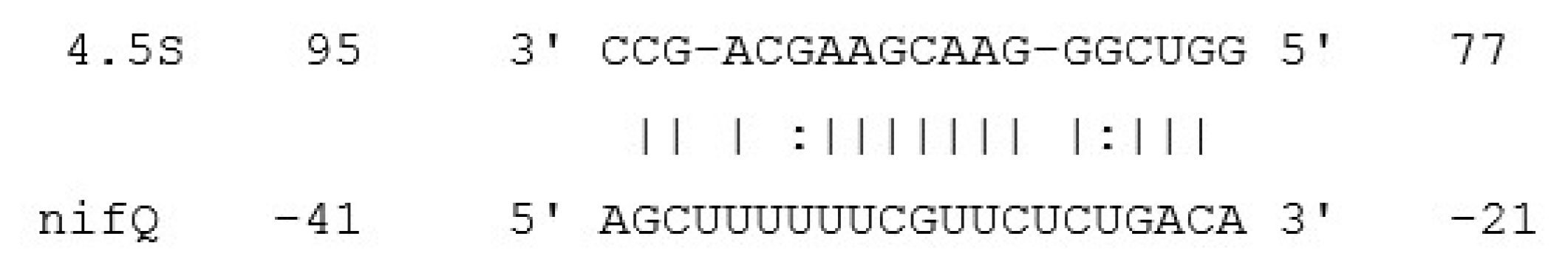

| Genus Herbaspirillum | ncRNAs |
| Herbaspirillum seropedicae Z67 | ohsC_RNA, Rhizobiales-2 and UPD-PKc |
| Herbaspirillum lusitanum P6-12 | Absence |
| Herbaspirillum hiltneri N3 | ar14, Acido-1, Alpha_RBS, ar14, asX2, asX3, Bp2_287, csRNA, isrN, istR, Ms_AS-4, Ms_IGR-8, ncr1175, NsiR1, P26, P5, PrrB_RsmZ, RsmY, RydC, sau-5971, SpF41_sRNA, Spot_42, Xoo5, sraA, sX9, TB10Cs4H2, TB11Cs5H2 and TB9Cs1H1 |
| Herbaspirillum frisingense GSF30 | Entero_5_CRE, P10 and STnc430 |
| Herbaspirillum spp. estirpe B65 | snoZ248 |
| Herbaspirillum rubrisubalbicans M1 | ALIL, Alpha_RBS, Archaea_SRP, asX2, b55, BjrC1505, C4, ceN84, istR, K_chan_RES, MAT2A_B, MEG3_2, NRF2_IRES, OrzO-P, P36, pRNA, PtaRNA1, rli60, rox2, rseX, sau-6072, sau-63, SpF25_sRNA, SpR10_sRNA, sroB, sX15, TB9Cs1H1 and TB9Cs1H2. |
| Herbaspirillum huttiense subsp. Putei estirpe IAM 15032 | asX2, Cardiovirus_CRE, Chloroflexi-1, MEG3_2, SBWMV2_UPD-PKk, sroB, veev_FSE, asX2, Cardiovirus_CRE, Chloroflexi-1, MEG3_2, SBWMV2_UPD-PKk, sroB and veev_FSE. |
| Herbaspirillum autotrophicum IAM 14942 | MIR1444 and SpF10_sRNA. |
| Herbaspirillum spp. estirpe B501 | Absence |
| Herbaspirillum spp. estirpe GW103 | sX2, Flavi_CRE, RyeB, SAM-IV, sau-6072 e sroB. |
| Herbaspirillum spp. estirpe RV1423 | psRNA2, sbcD and STnc40. |
| Herbaspirillum rhizosphaerae UMS-37 | NRF2_IRES, NsiR1, Pseudomon-groES and sX9. |
| Herbaspirillum rubrisubalbicans spp. estirpe Os34 | ryfA, SpR20_sRNA and asX2. |
| Herbaspirillum rubrisubalbicans spp. estirpe Os45 | asX2, ROSE_2 and ryfA. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carvalho Garcia, A.; Dos Santos, V.L.P.; Santos Cavalcanti, T.C.; Collaço, L.M.; Graf, H. Bacterial Small RNAs in the Genus Herbaspirillum spp. Int. J. Mol. Sci. 2019, 20, 46. https://doi.org/10.3390/ijms20010046
Carvalho Garcia A, Dos Santos VLP, Santos Cavalcanti TC, Collaço LM, Graf H. Bacterial Small RNAs in the Genus Herbaspirillum spp. International Journal of Molecular Sciences. 2019; 20(1):46. https://doi.org/10.3390/ijms20010046
Chicago/Turabian StyleCarvalho Garcia, Amanda, Vera Lúcia Pereira Dos Santos, Teresa Cristina Santos Cavalcanti, Luiz Martins Collaço, and Hans Graf. 2019. "Bacterial Small RNAs in the Genus Herbaspirillum spp." International Journal of Molecular Sciences 20, no. 1: 46. https://doi.org/10.3390/ijms20010046
APA StyleCarvalho Garcia, A., Dos Santos, V. L. P., Santos Cavalcanti, T. C., Collaço, L. M., & Graf, H. (2019). Bacterial Small RNAs in the Genus Herbaspirillum spp. International Journal of Molecular Sciences, 20(1), 46. https://doi.org/10.3390/ijms20010046






