Proteomic Analysis of the Breast Cancer Brain Metastasis Microenvironment
Abstract
:1. Introduction
2. Results
2.1. Proteomic Profiling of the Brain Microenvironment in Experimental Brain Metastases
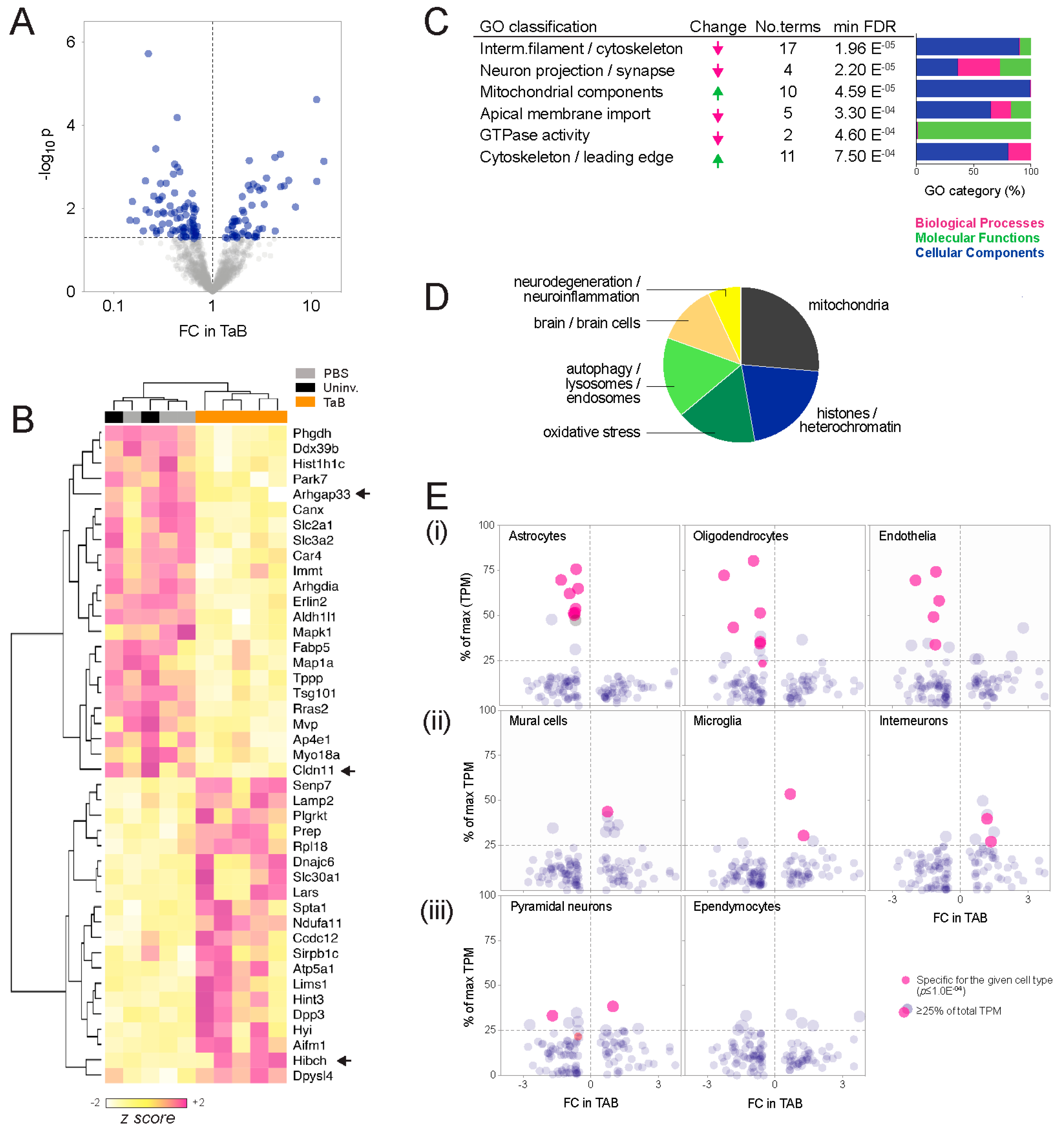
- Ontology enrichment analysis. Analysis of interconnectivity amongst the differential proteins using the STRING database [14] showed significant enrichment of known protein-protein interactions, implying biological relatedness (high-confidence interactions p = 3.4 × 10−8; Figure S1). To investigate whether these networks represent particular bio-ontologies, we performed functional enrichment analysis and consolidated the associated ontology terms according to semantic similarity using REVIGO [15]. This indicated that components, processes and functions typically associated with neurons decreased in graft-associated brain tissue, and those associated with metabolic reprogramming or cell migration increased (Figure 2C; Table S2). Cellular components (CC, compartments or stable macromolecular complexes) were the predominating category, and construction of a similarity network from the CC terms revealed two major clusters: One comprising mitochondria and vesicular elements, and a second containing mainly cytoskeletal and neural projection elements (Figure S2; Table S2). Consistent with this, high-throughput PubMed text mining indicated that the differentially abundant proteins have been most frequently studied in the contexts of mitochondria, histones, oxidative stress and autophagy (Figure 2D; Table S3).
- Cell composition analysis. The proteomic landscape of graft-associated brain tissue could also reflect changes in the relative abundance of different cell types, so we investigated this using meta-analysis of single-cell RNA sequencing (scRNAseq) data generated by Zeisel and colleagues from pooled cortical and hippocampal mouse brain tissue [16]. While based on transcriptomic rather than proteomic output, it is the largest (n = 3005 cells) and most comprehensive expression mouse brain cell dataset available. Considering the proteomic data as a survey of tissue composition, we compared the changes observed in the SWATH-MS experiment with cell type specificity at the RNA level, defined by statistical association (ANOVA test; p ≤ 1.0 × 10−4), as well as the proportion expressed by each cell type (Figure 2E). This established glial markers (e.g., Claudin-11 (oligodendrocyte-specific protein; OSP) and Aldh1l1 (10-formyltetra-hydrofolate dehydrogenase), associated with astrocytes and oligodendrocytes, respectively [17,18,19,20,21]), confirming the meta-analysis as a valid way to identify cell type-associated transcripts. Overall, transcripts associated with astrocytes, oligodendrocytes and endothelia were less abundant at the protein level in TAB, while those associated with mural cells (pericytes and vascular smooth muscle), microglia and interneurons were more abundant.
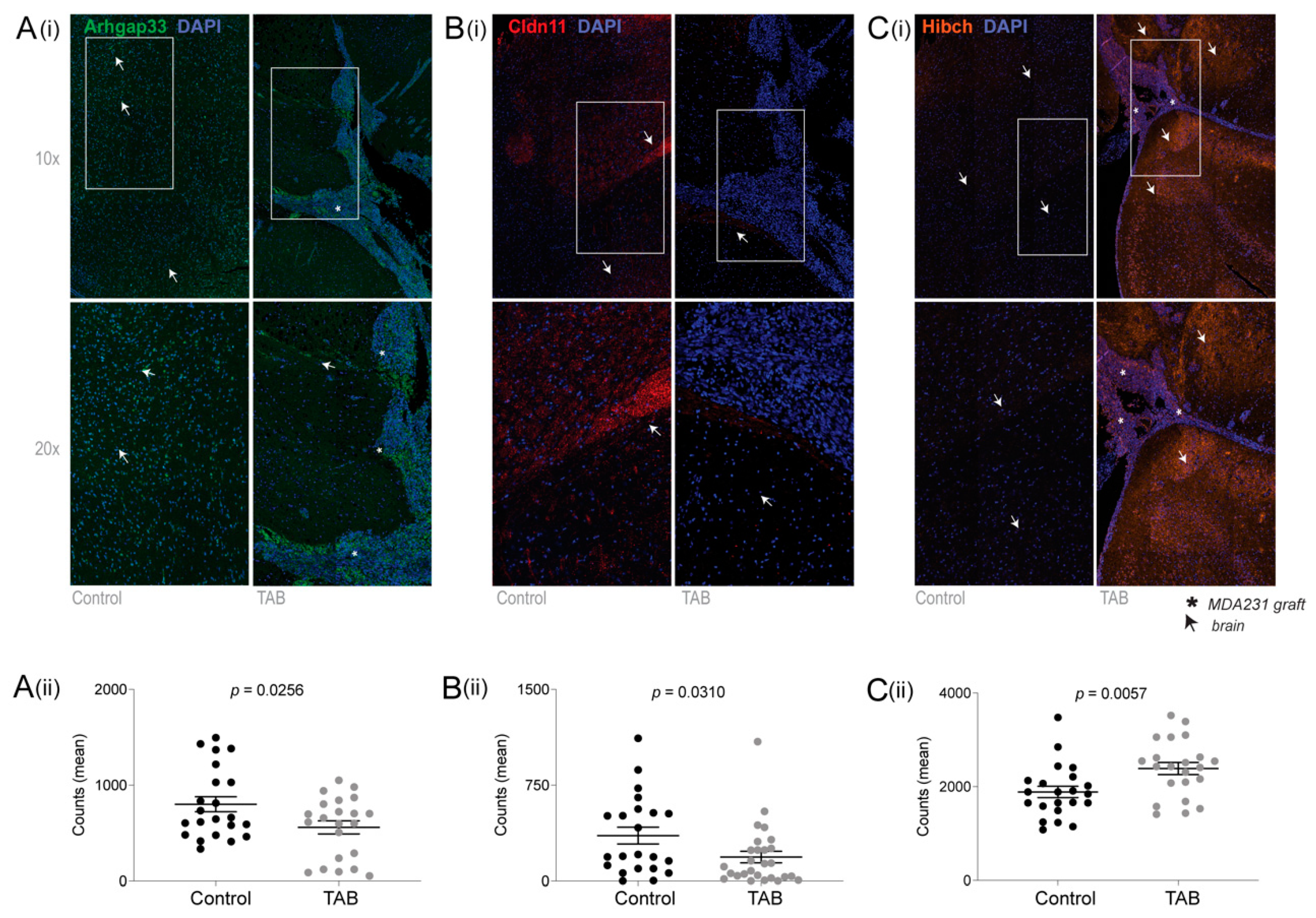
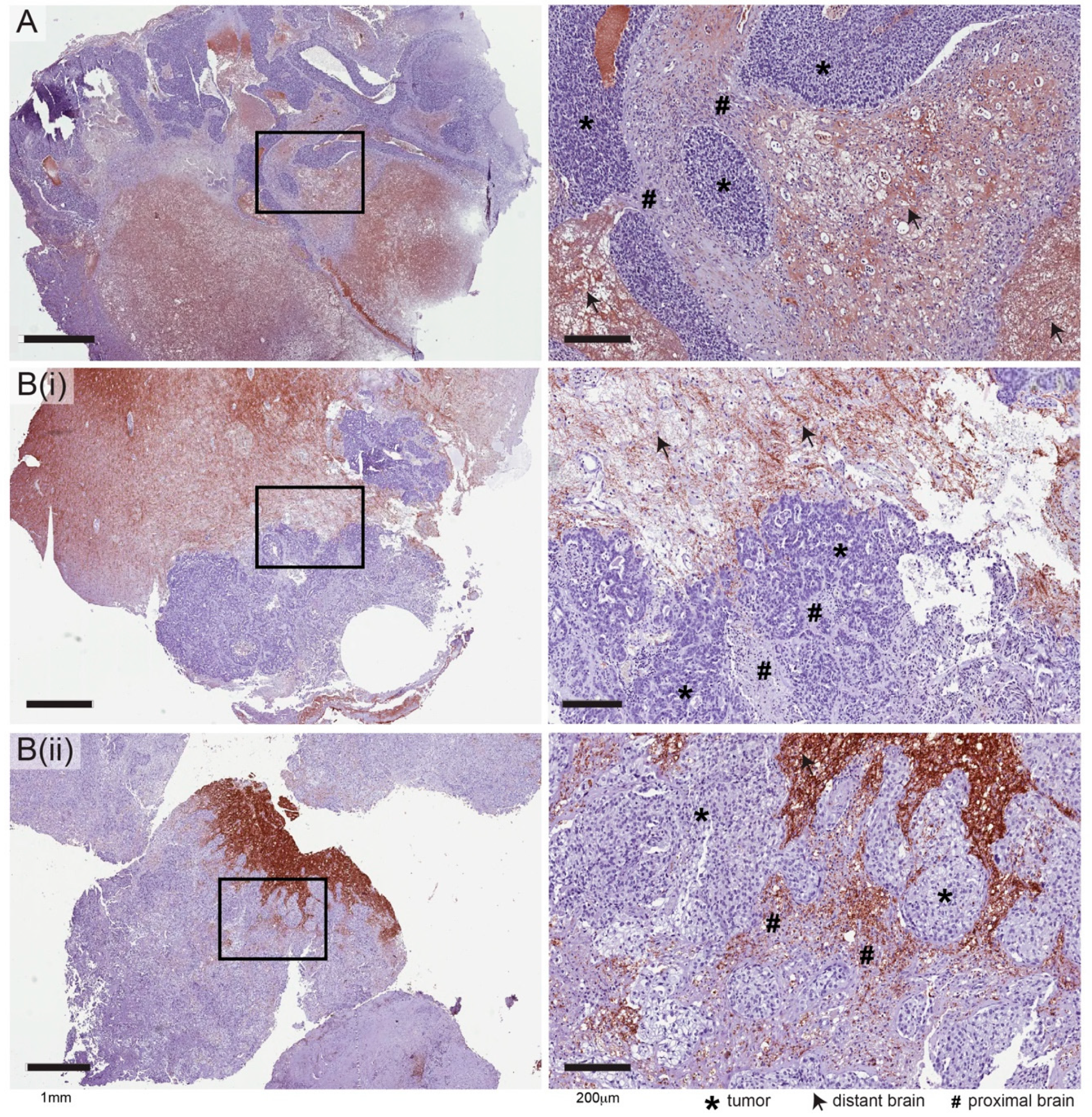
2.2. Independent Validation of HIBCH, Cldn11 and Arhgap33
2.3. Expression of HIBCH, CLDN11 and ARHGAP33 Proteins in Human Brain Metastases
3. Discussion
4. Materials and Methods
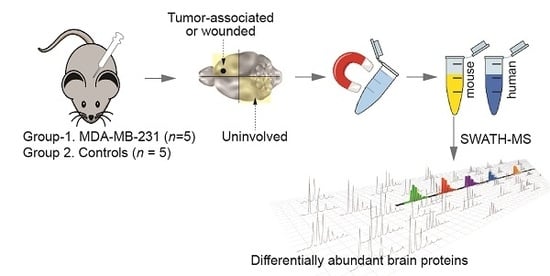
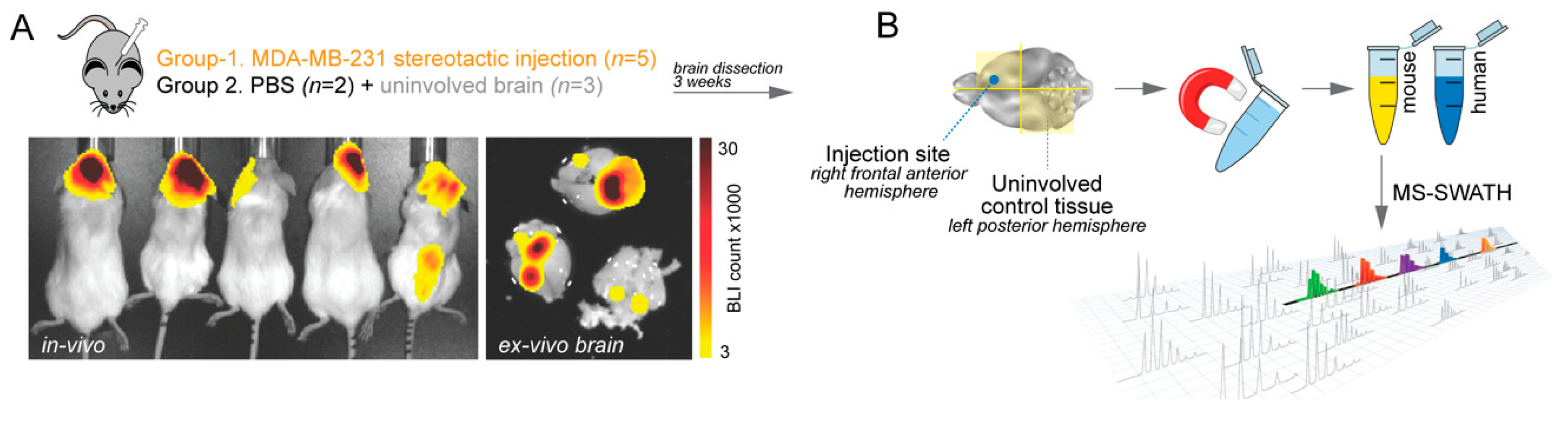
4.1. Experimental Brain Metastases for Tumour Microenvironment Profiling
4.2. Cell Separation
4.3. Sample Preparation and Mass Spectrometry (MS)
4.4. Identification of Differentially Abundant Proteins
4.5. Bioinformatics
4.6. Fluorescent Multiplex Immunohistochemistry (fmIHC) and Multispectral Imaging
4.7. Immunohistochemistry (IHC) Analysis of Craniotomy Specimens
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| FFPE | Formalin fixed paraffin embedded |
| HIBCH | 3-hydroxyisobutyryl-CoA hydrolase |
| Cldn11 | Claudin 11 |
| TSA | Tyramide signal amplification |
| GFAP | Glial fibrillary acidic protein |
| BSA | Bovine serum albumin |
| RT | Room temperature |
References
- Tabouret, E.; Chinot, O.; Metellus, P.; Tallet, A.; Viens, P.; Goncalves, A. Recent trends in epidemiology of brain metastases: An overview. Anticancer Res. 2012, 32, 4655–4662. [Google Scholar] [PubMed]
- Bos, P.D.; Zhang, X.H.F.; Nadal, C.; Shu, W.; Gomis, R.R.; Nguyen, D.X.; Minn, A.J.; van de Vijver, M.J.; Gerald, W.L.; Foekens, J.A.; et al. Genes that mediate breast cancer metastasis to the brain. Nature 2009, 459, 1005–1009. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, L.; Simpson, P.T.; Smart, C.E.; Cocciardi, S.; Waddell, N.; Lane, A.; Morrison, B.J.; Vargas, A.; Healey, S.; Beesley, J.; et al. HER3 and downstream pathways are involved in colonization of brain metastases from breast cancer. Breast Cancer Res. 2010, 12, R46. [Google Scholar] [CrossRef]
- Li, F.; Glinskii, O.V.; Zhou, J.; Wilson, L.S.; Barnes, S.; Anthony, D.C.; Glinsky, V.V. Identification and analysis of signaling networks potentially involved in breast carcinoma metastasis to the brain. PLoS ONE 2011, 6, e21977. [Google Scholar] [CrossRef]
- Hohensee, I.; Lamszus, K.; Riethdorf, S.; Meyer-Staeckling, S.; Glatzel, M.; Matschke, J.; Witzel, I.; Westphal, M.; Brandt, B.; Muller, V.; et al. Frequent genetic alterations in EGFR- and HER2-driven pathways in breast cancer brain metastases. Am. J. Pathol. 2013, 183, 83–95. [Google Scholar] [CrossRef]
- Dun, M.D.; Chalkley, R.J.; Faulkner, S.; Keene, S.; Avery-Kiejda, K.A.; Scott, R.J.; Falkenby, L.G.; Cairns, M.J.; Larsen, M.R.; Bradshaw, R.A.; et al. Proteotranscriptomic Profiling of 231-BR Breast Cancer Cells: Identification of Potential Biomarkers and Therapeutic Targets for Brain Metastasis. Mol. Cell. Proteomics 2015, 14, 2316–2330. [Google Scholar] [CrossRef] [Green Version]
- Valiente, M.; Obenauf, A.C.; Jin, X.; Chen, Q.; Zhang, X.H.; Lee, D.J.; Chaft, J.E.; Kris, M.G.; Huse, J.T.; Brogi, E.; et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell 2014, 156, 1002–1016. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. The Microenvironmental Landscape of Brain Tumors. Cancer Cell 2017, 31, 326–341. [Google Scholar] [CrossRef] [Green Version]
- Gril, B.; Palmieri, D.; Qian, Y.; Anwar, T.; Liewehr, D.J.; Steinberg, S.M.; Andreu, Z.; Masana, D.; Fernandez, P.; Steeg, P.S.; et al. Pazopanib inhibits the activation of PDGFRbeta-expressing astrocytes in the brain metastatic microenvironment of breast cancer cells. Am. J. Pathol. 2013, 182, 2368–2379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.J.; Zhang, Q.; Huang, W.C.; Li, P.; Li, M.; Wang, X.; Zhang, C.; et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015, 527, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Boire, A.; Jin, X.; Valiente, M.; Er, E.E.; Lopez-Soto, A.; Jacob, L.S.; Patwa, R.; Shah, H.; Xu, K.; et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 2016, 533, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Balasubramanian, K.; Fan, D.; Kim, S.J.; Guo, L.; Wang, H.; Bar-Eli, M.; Aldape, K.D.; Fidler, I.J. Reactive astrocytes protect melanoma cells from chemotherapy by sequestering intracellular calcium through gap junction communication channels. Neoplasia 2010, 12, 748–754. [Google Scholar] [CrossRef]
- Arvanitis, C.D.; Askoxylakis, V.; Guo, Y.; Datta, M.; Kloepper, J.; Ferraro, G.B.; Bernabeu, M.O.; Fukumura, D.; McDannold, N.; Jain, R.K. Mechanisms of enhanced drug delivery in brain metastases with focused ultrasound-induced blood-tumor barrier disruption. PNAS 2018, 115, E8717–E8726. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Zeisel, A.; Munoz-Manchado, A.B.; Codeluppi, S.; Lonnerberg, P.; La Manno, G.; Jureus, A.; Marques, S.; Munguba, H.; He, L.; Betsholtz, C.; et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 2015, 347, 1138–1142. [Google Scholar] [CrossRef]
- Park, Y.; Liu, C.; Luo, T.; Dietrich, W.D.; Bramlett, H.; Hu, B. Chaperone-Mediated Autophagy after Traumatic Brain Injury. J. Neurotrauma 2015, 32, 1449–1457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, H.K.; Ji, K.M.; Kim, J.; Jou, I.; Joe, E.H. Repair of astrocytes, blood vessels, and myelin in the injured brain: Possible roles of blood monocytes. Mol. Brain 2013, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Bronstein, J.M.; Tiwari-Woodruff, S.; Buznikov, A.G.; Stevens, D.B. Involvement of OSP/claudin-11 in oligodendrocyte membrane interactions: Role in biology and disease. J. Neurosci. Res. 2000, 59, 706–711. [Google Scholar] [CrossRef]
- Anthony, T.E.; Heintz, N. The folate metabolic enzyme ALDH1L1 is restricted to the midline of the early CNS, suggesting a role in human neural tube defects. J. Comp. Neurol. 2007, 500, 368–383. [Google Scholar] [CrossRef]
- John Lin, C.C.; Yu, K.; Hatcher, A.; Huang, T.W.; Lee, H.K.; Carlson, J.; Weston, M.C.; Chen, F.; Zhang, Y.; Zhu, W.; et al. Identification of diverse astrocyte populations and their malignant analogs. Nat. Neurosci. 2017, 20, 396–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.; Ha, C.M.; Chang, S. SNX26, a GTPase-activating protein for Cdc42, interacts with PSD-95 protein and is involved in activity-dependent dendritic spine formation in mature neurons. J. Biol. Chem. 2013, 288, 29453–29466. [Google Scholar] [CrossRef]
- Nakazawa, T.; Hashimoto, R.; Sakoori, K.; Sugaya, Y.; Tanimura, A.; Hashimotodani, Y.; Ohi, K.; Yamamori, H.; Yasuda, Y.; Umeda-Yano, S.; et al. Emerging roles of ARHGAP33 in intracellular trafficking of TrkB and pathophysiology of neuropsychiatric disorders. Nat. Commun. 2016, 7, 10594. [Google Scholar] [CrossRef]
- Denninger, A.R.; Breglio, A.; Maheras, K.J.; LeDuc, G.; Cristiglio, V.; Deme, B.; Gow, A.; Kirschner, D.A. Claudin-11 Tight Junctions in Myelin Are a Barrier to Diffusion and Lack Strong Adhesive Properties. Biophys. J. 2015, 109, 1387–1397. [Google Scholar] [CrossRef] [Green Version]
- Lim, M.; Puttick, S.; Houston, Z.H.; Thurecht, K.J.; Kalita-de Croft, P.; Mahler, S.; Rose, S.E.; Jeffree, R.L.; Mazzieri, R.; Dolcetti, R.; et al. Innovative Therapeutic Strategies for Effective Treatment of Brain Metastases. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- McCart Reed, A.E.; Saunus, J.M.; Ferguson, K.; Niland, C.; Simpson, P.T.; Lakhani, S.R. The Brisbane Breast Bank. Open J. Bioresour. 2018, 5, 5. [Google Scholar] [CrossRef]
- Charles, N.A.; Holland, E.C.; Gilbertson, R.; Glass, R.; Kettenmann, H. The brain tumor microenvironment. Glia 2012, 60, 502–514. [Google Scholar] [CrossRef]
- Sevenich, L.; Bowman, R.L.; Mason, S.D.; Quail, D.F.; Rapaport, F.; Elie, B.T.; Brogi, E.; Brastianos, P.K.; Hahn, W.C.; Holsinger, L.J.; et al. Analysis of tumour- and stroma-supplied proteolytic networks reveals a brain-metastasis-promoting role for cathepsin S. Nat. Cell Biol. 2014, 16, 876–888. [Google Scholar] [CrossRef] [Green Version]
- Sartorius, C.A.; Hanna, C.T.; Gril, B.; Cruz, H.; Serkova, N.J.; Huber, K.M.; Kabos, P.; Schedin, T.B.; Borges, V.F.; Steeg, P.S.; et al. Estrogen promotes the brain metastatic colonization of triple negative breast cancer cells via an astrocyte-mediated paracrine mechanism. Oncogene 2016, 35, 2881–2892. [Google Scholar] [CrossRef] [PubMed]
- Lyle, L.T.; Lockman, P.R.; Adkins, C.E.; Mohammad, A.S.; Sechrest, E.; Hua, E.; Palmieri, D.; Liewehr, D.J.; Steinberg, S.M.; Kloc, W.; et al. Alterations in Pericyte Subpopulations Are Associated with Elevated Blood-Tumor Barrier Permeability in Experimental Brain Metastasis of Breast Cancer. Clin. Cancer Res. 2016, 22, 5287–5299. [Google Scholar] [CrossRef] [PubMed]
- Burda, J.E.; Sofroniew, M.V. Reactive gliosis and the multicellular response to CNS damage and disease. Neuron 2014, 81, 229–248. [Google Scholar] [CrossRef] [PubMed]
- Sperringer, J.E.; Addington, A.; Hutson, S.M. Branched-Chain Amino Acids and Brain Metabolism. Neurochem. Res. 2017, 42, 1697–1709. [Google Scholar] [CrossRef]
- Shimomura, Y.; Murakami, T.; Fujitsuka, N.; Nakai, N.; Sato, Y.; Sugiyama, S.; Shimomura, N.; Irwin, J.; Hawes, J.W.; Harris, R.A. Purification and partial characterization of 3-hydroxyisobutyryl-coenzyme A hydrolase of rat liver. J. Biol. Chem. 1994, 269, 14248–14253. [Google Scholar]
- Dupont, N.; Orhon, I.; Bauvy, C.; Codogno, P. Autophagy and autophagic flux in tumor cells. Methods Enzymol. 2014, 543, 73–88. [Google Scholar] [CrossRef]
- Meijer, A.J.; Lorin, S.; Blommaart, E.F.; Codogno, P. Regulation of autophagy by amino acids and MTOR-dependent signal transduction. Amino Acids 2015, 47, 2037–2063. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lee, H.J.; Wu, X.; Huo, L.; Kim, S.J.; Xu, L.; Wang, Y.; He, J.; Bollu, L.R.; Gao, G.; et al. Gain of glucose-independent growth upon metastasis of breast cancer cells to the brain. Cancer Res. 2015, 75, 554–565. [Google Scholar] [CrossRef]
- Caruso Bavisotto, C.; Scalia, F.; Marino Gammazza, A.; Carlisi, D.; Bucchieri, F.; Conway de Macario, E.; Macario, A.J.L.; Cappello, F.; Campanella, C. Extracellular Vesicle-Mediated Cell(-)Cell Communication in the Nervous System: Focus on Neurological Diseases. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Doron, H.; Pukrop, T.; Erez, N. A Blazing Landscape: Neuroinflammation Shapes Brain Metastasis. Cancer Res. 2019, 79, 423–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.X.; Song, X.; Pascovici, D.; Zaw, T.; Care, N.; Krisp, C.; Molloy, M.P. SWATH Mass Spectrometry Performance Using Extended Peptide MS/MS Assay Libraries. Mol. Cell. Proteomics 2016, 15, 2501–2514. [Google Scholar] [CrossRef] [Green Version]
- UniProt Consortium, T. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalita-de Croft, P.; Straube, J.; Lim, M.; Al-Ejeh, F.; Lakhani, S.R.; Saunus, J.M. Proteomic Analysis of the Breast Cancer Brain Metastasis Microenvironment. Int. J. Mol. Sci. 2019, 20, 2524. https://doi.org/10.3390/ijms20102524
Kalita-de Croft P, Straube J, Lim M, Al-Ejeh F, Lakhani SR, Saunus JM. Proteomic Analysis of the Breast Cancer Brain Metastasis Microenvironment. International Journal of Molecular Sciences. 2019; 20(10):2524. https://doi.org/10.3390/ijms20102524
Chicago/Turabian StyleKalita-de Croft, Priyakshi, Jasmin Straube, Malcolm Lim, Fares Al-Ejeh, Sunil R. Lakhani, and Jodi M. Saunus. 2019. "Proteomic Analysis of the Breast Cancer Brain Metastasis Microenvironment" International Journal of Molecular Sciences 20, no. 10: 2524. https://doi.org/10.3390/ijms20102524