DRL1, Encoding A NAC Transcription Factor, Is Involved in Leaf Senescence in Grapevine
Abstract
:1. Introduction
2. Results
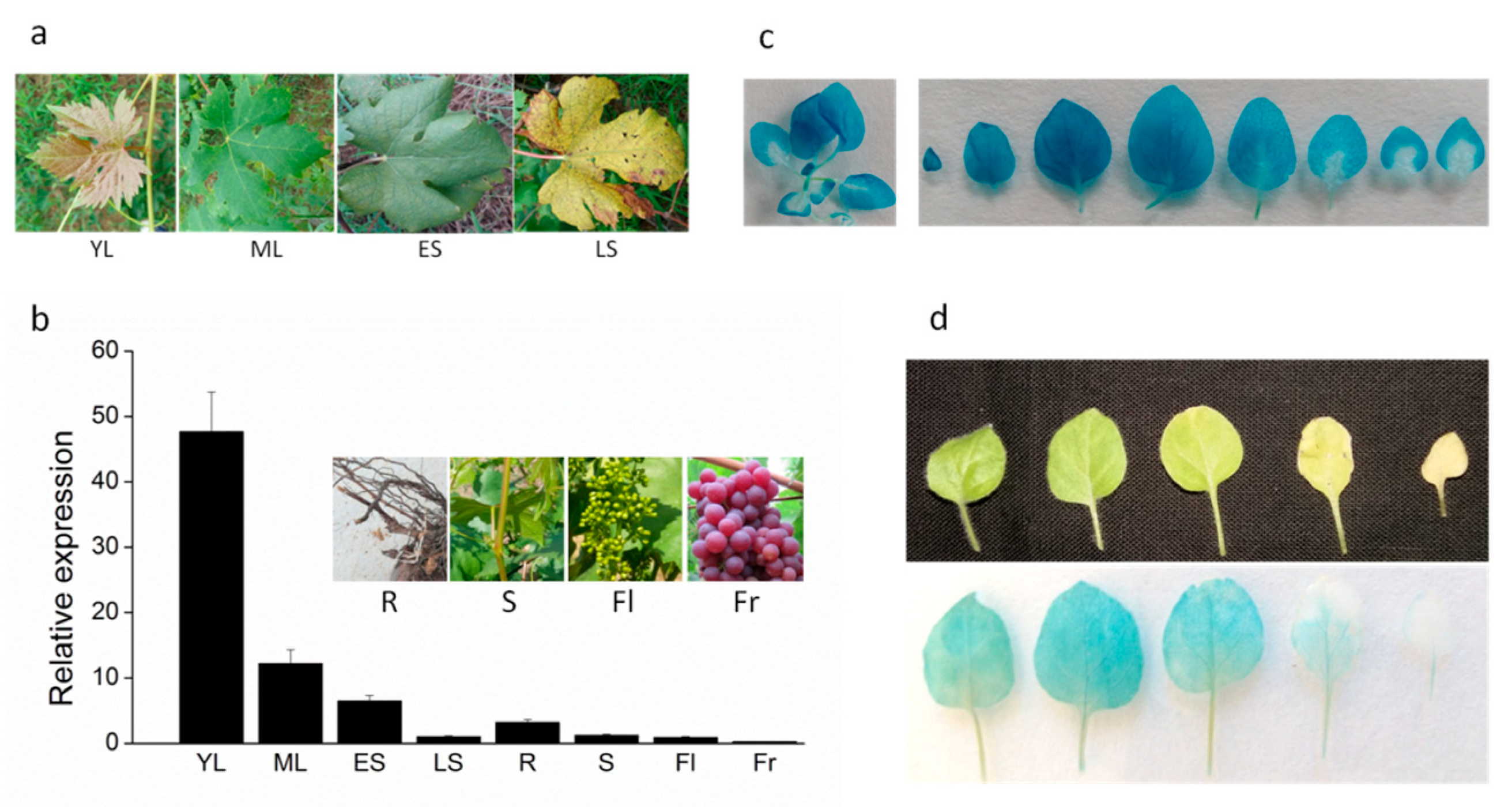
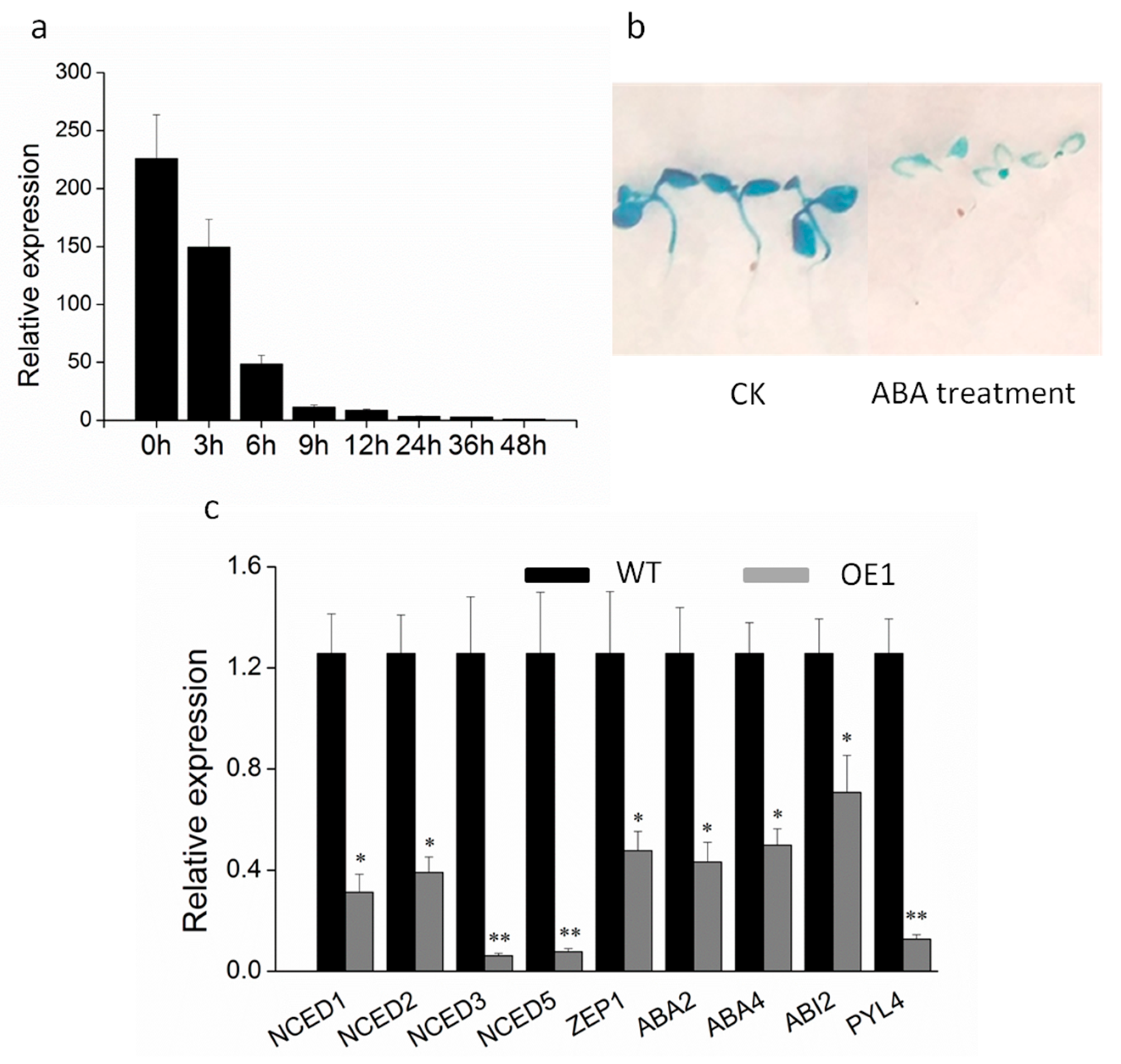
2.1. DRL1 is Down-Regulated during Leaf Senescence in Grapevine
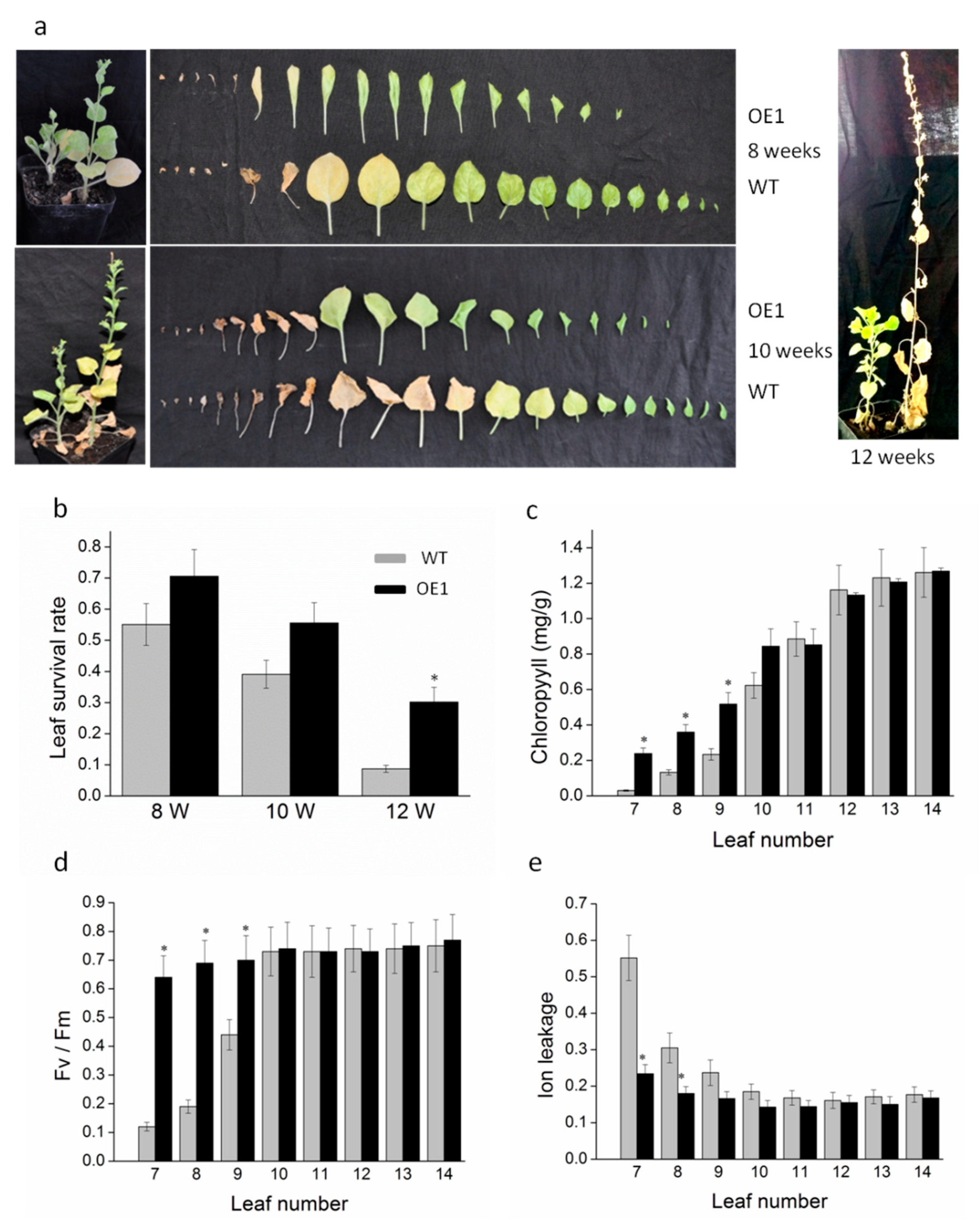
2.2. Ectopic Overexpression of DRL1 Delayed Leaf Senescence
2.3. Analysis of Differentially Expressed Genes (DEGs) in DRL1-Overexpressing Transgenic Plants
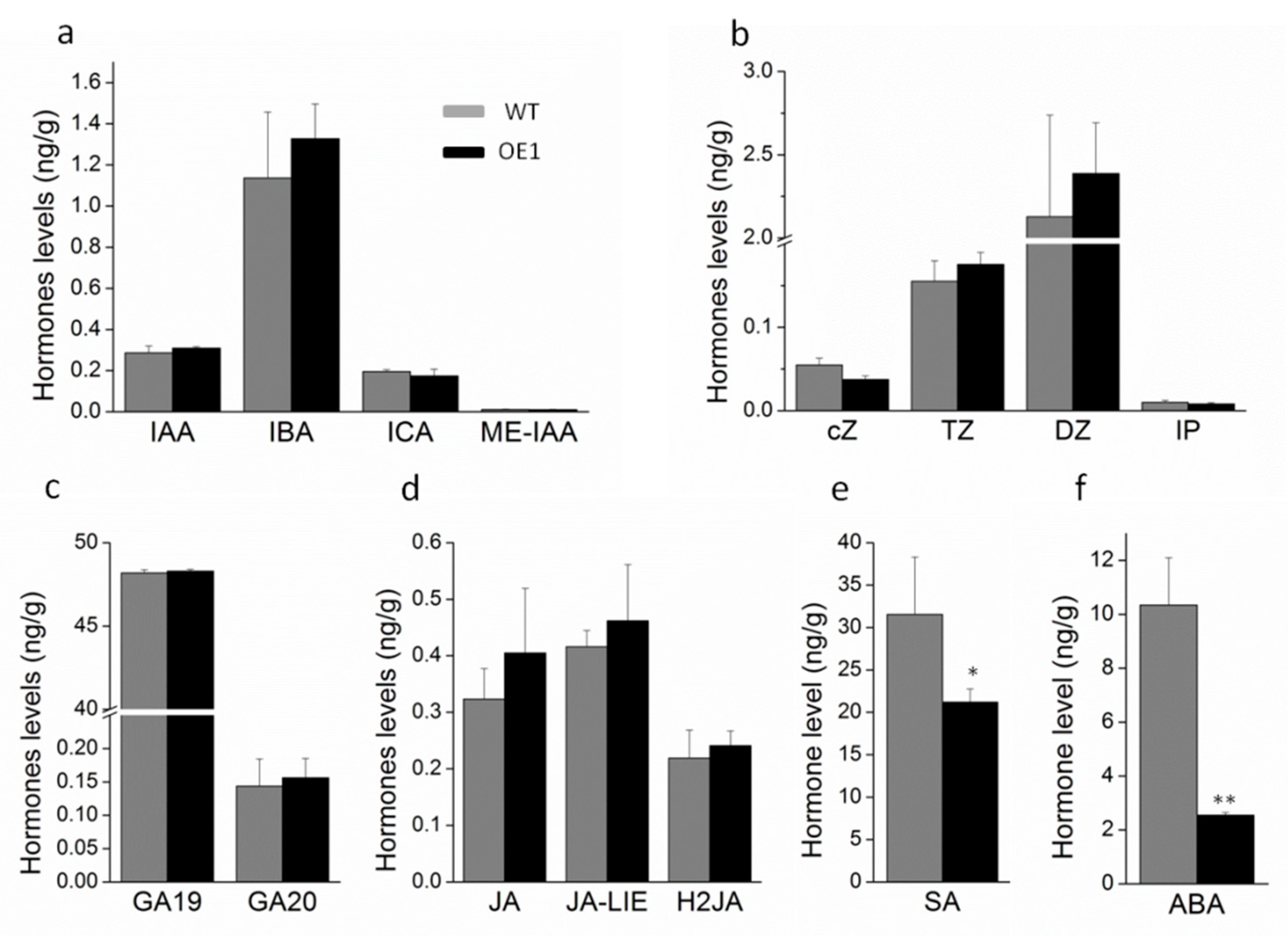
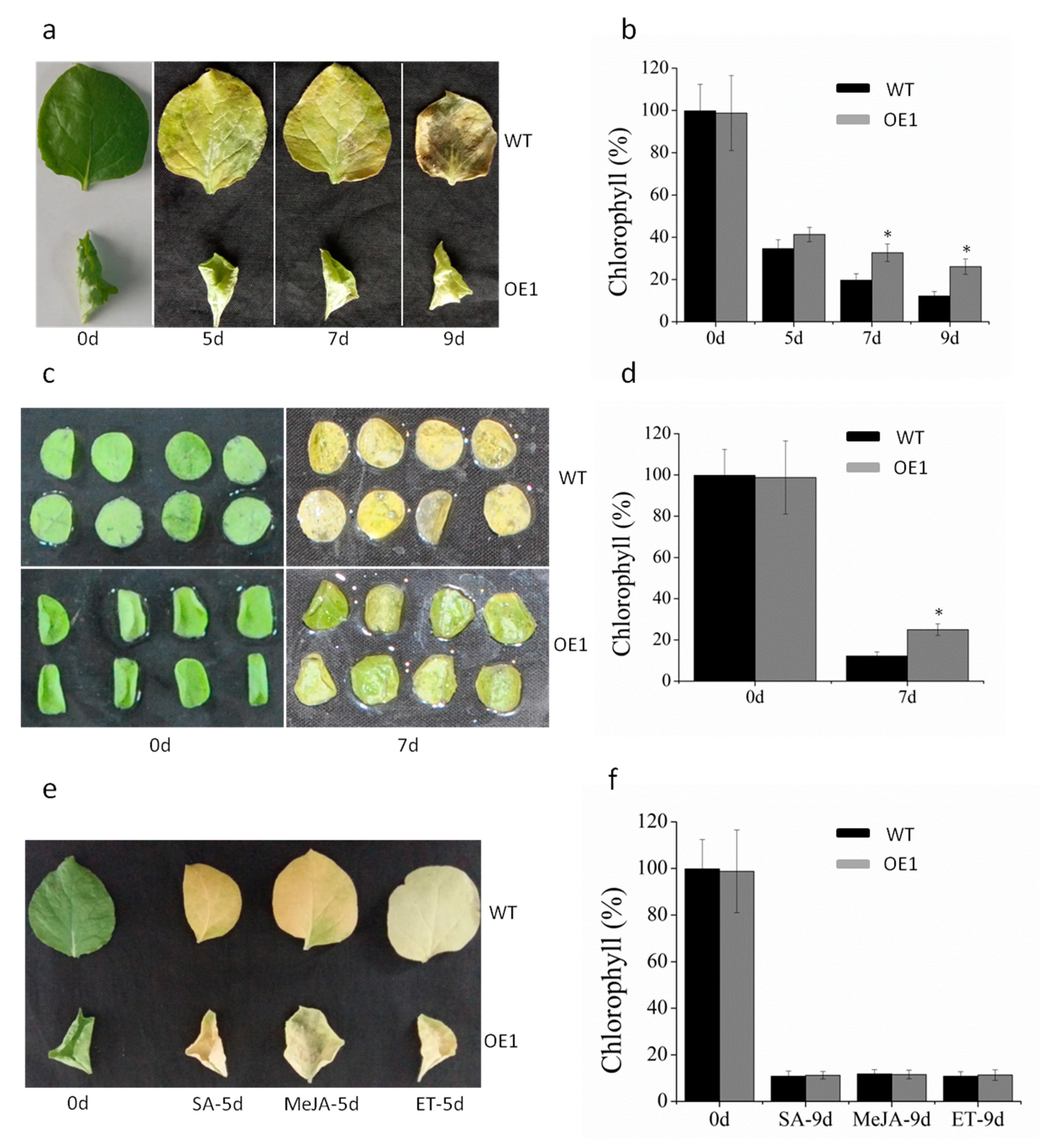
2.4. ABA Is Involved in Leaf Senescence of DRL1-Overexpressing Transgenic Plants
2.5. DRL1 Delayed Leaf Senescence by Regulation of ABA
3. Discussion
4. Materials and Methods
4.1. Plant Material and Hormone Treatment
4.2. RNA Analysis
4.3. Production of Transgenic Tobacco
4.4. Chlorophyll Concentration, Fluorescence, and Ion Leakage Analysis
4.5. Transcriptome Analysis
4.6. Hormones Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| Chl | Chlorophyll |
| CTAB | Cetyltrimethylammonium bromide |
| DEGs | Differentially expressed genes |
| GUS | β-glucuronidase |
| JA | Jasmonic acid |
| NCED | 9-cisepoxycarotenoid dioxygenase |
| SA | Salicylic acid |
| ZEP | Zeaxanthin epoxidase |
References
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S. Leaf senescence: Signals, execution, and regulation. Curr. Top Dev. Biol. 2005, 71, 83–112. [Google Scholar] [PubMed]
- Kant, S.; Burch, D.; Badenhorst, P.; Palanisamy, R.; Mason, J.; Spangenberg, G. Regulated expression of a cytokinin biosynthesis gene IPT delays leaf senescence and improves yield under rainfed and irrigated conditions in canola (Brassica napus L.). PLoS ONE 2015, 10, e0116349. [Google Scholar] [CrossRef]
- Robson, P.R.; Donnison, I.S.; Wang, K.; Frame, B.; Pegg, S.E.; Thomas, A.; Thomas, H. Leaf senescence is delayed in maize expressing the Agrobacterium IPT gene under the control of a novel maize senescence-enhanced promoter. Plant Biotechnol. J. 2004, 2, 101–112. [Google Scholar] [CrossRef] [Green Version]
- Van der Graaff, E.; Schwacke, R.; Schneider, A.; Desimone, M.; Flugge, U.I.; Kunze, R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 2006, 141, 776–792. [Google Scholar] [CrossRef]
- Jibran, R.; Hunter, D.A.; Dijkwel, P.P. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 2013, 82, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Osborne, D.J. Hormonal regulation of leaf senescence. Symp. Soc. Exp. Biol. 1967, 21, 305–321. [Google Scholar] [PubMed]
- He, Y.; Fukushige, H.; Hildebrand, D.F.; Gan, S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Guo, H. Ethylene Treatment in studying leaf senescence in Arabidopsis. Methods Mol. Biol. 2018, 1744, 105–112. [Google Scholar]
- Qiu, K.; Li, Z.; Yang, Z.; Chen, J.; Wu, S.; Zhu, X.; Gao, S.; Gao, J.; Ren, G.; Kuai, B.; et al. EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet 2015, 11, e1005399. [Google Scholar] [CrossRef]
- Li, Z.; Peng, J.; Wen, X.; Guo, H. Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 2013, 25, 3311–3328. [Google Scholar] [CrossRef]
- Lee, S.H.; Sakuraba, Y.; Lee, T.; Kim, K.W.; An, G.; Lee, H.Y.; Paek, N.C. Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J. Integr. Plant Biol. 2015, 57, 562–576. [Google Scholar] [CrossRef]
- Morris, K.; MacKerness, S.A.; Page, T.; John, C.F.; Murphy, A.M.; Carr, J.P.; Buchanan-Wollaston, V. Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 2000, 23, 677–685. [Google Scholar] [CrossRef]
- Quattrocchio, F.; Wing, J.; van der Woude, K.; Souer, E.; de Vetten, N.; Mol, J.; Koes, R. Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 1999, 11, 1433–1444. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Souer, E.; vanHouwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef]
- Hu, R.; Qi, G.; Kong, Y.; Kong, D.; Gao, Q.; Zhou, G. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 2010, 10, 145. [Google Scholar] [CrossRef]
- Li, C.W.; Su, R.C.; Cheng, C.P.; Sanjaya; You, S.J.; Hsieh, T.H.; Chao, T.C.; Chan, M.T. Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway. Plant Physiol. 2011, 156, 213–227. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Oh, S.K.; Lee, S.; Yu, S.; Choi, D. Expression of a novel NAC domain-containing transcription factor (CaNAC1) is preferentially associated with incompatible interactions between chili pepper and pathogens. Planta 2005, 222, 876–887. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef]
- Zhao, D.; Derkx, A.P.; Liu, D.C.; Buchner, P.; Hawkesford, M.J. Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol. 2015, 17, 904–913. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Qiu, K.; Kuai, B.; Ding, Y. Identification of an NAP-like transcription factor BeNAC1 regulating leaf senescence in bamboo (Bambusa emeiensis “Viridiflavus”). Physiol. Plant 2011, 142, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [PubMed]
- Wang, N.; Zheng, Y.; Xin, H.; Fang, L.; Li, S. Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep. 2013, 32, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Le Henanff, G.; Profizi, C.; Courteaux, B.; Rabenoelina, F.; Gerard, C.; Clement, C.; Baillieul, F.; Cordelier, S.; Dhondt-Cordelier, S. Grapevine NAC1 transcription factor as a convergent node in developmental processes, abiotic stresses, and necrotrophic/biotrophic pathogen tolerance. J. Exp. Bot. 2013, 64, 4877–4893. [Google Scholar] [CrossRef]
- Fang, L.; Su, L.; Sun, X.; Li, X.; Sun, M.; Karungo, S.K.; Fang, S.; Chu, J.; Li, S.; Xin, H. Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synthesis. J. Exp. Bot. 2016, 67, 2829–2845. [Google Scholar] [CrossRef] [PubMed]
- Tello, J.; Torres-Perez, R.; Grimplet, J.; Carbonell-Bejerano, P.; Martinez-Zapater, J.M.; Ibanez, J. Polymorphisms and minihaplotypes in the VvNAC26 gene associate with berry size variation in grapevine. BMC Plant Biol. 2015, 15, 253. [Google Scholar] [CrossRef]
- Yan, C.; Li, G.; Mu, J.; Lou, H.; Zhu, Z. Subcellular localization and functional analysis of a NAC gene VvDRL1 from Vitis vinifera “Yatomi Rosa”. Acta Horticulturae Sinica 2016, 43, 643–652. [Google Scholar]
- Diago, M.P.; Correa, C.; Millan, B.; Barreiro, P.; Valero, C.; Tardaguila, J. Grapevine yield and leaf area estimation using supervised classification methodology on RGB images taken under field conditions. Sensors 2012, 12, 16988–17006. [Google Scholar] [CrossRef]
- Wu, X.Y.; Kuai, B.K.; Jia, J.Z.; Jing, H.C. Regulation of leaf senescence and crop genetic improvement. J. Integ. Plant Biol. 2012, 54, 936–952. [Google Scholar] [CrossRef]
- Antonello, J.M.; Clark, R.L.; Heyse, J.F. Application of the tukey trend test procedure to assess developmental and reproductive toxicity. I. Measurement data. Fundam. Appl. Toxicol. 1993, 21, 52–58. [Google Scholar] [CrossRef]
- Moulia-Pelat, J.P.; Spiegel, A.; Baudon, D.; Merlin, M.; Lemardeley, P.; Daumerie, D.; Gateff, C. Epidemiological value of graphic representation of the Tukey test. Med. Trop. (Mars) 1989, 49, 285–288. [Google Scholar]
- Zhou, Y.; Huang, W.; Liu, L.; Chen, T.; Zhou, F.; Lin, Y. Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol. 2013, 13, 132. [Google Scholar] [CrossRef]
- Hou, K.; Wu, W.; Gan, S.S. SAUR36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiol. 2013, 161, 1002–1009. [Google Scholar] [CrossRef]
- Gan, S.; Amasino, R.M. Inhibition of leaf senescence by autoregulated production of cytokinin. Science 1995, 270, 1986–1988. [Google Scholar] [CrossRef]
- He, Y.; Gan, S. A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell 2002, 14, 805–815. [Google Scholar] [CrossRef]
- Gregersen, P.L.; Holm, P.B. Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biotechnol. J. 2007, 5, 192–206. [Google Scholar] [CrossRef]
- Balazadeh, S.; Riano-Pachon, D.M.; Mueller-Roeber, B. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol. (Stuttg) 2008, 10 Suppl. 1, 63–75. [Google Scholar] [CrossRef]
- Guo, Y.; Cai, Z.; Gan, S. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 2004, 27, 521–549. [Google Scholar] [CrossRef]
- Shah, S.T.; Pang, C.; Fan, S.; Song, M.; Arain, S.; Yu, S. Isolation and expression profiling of GhNAC transcription factor genes in cotton (Gossypium hirsutum L.) during leaf senescence and in response to stresses. Gene 2013, 531, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC Gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Seo, P.J.; Lee, H.J.; Park, C.M. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 2012, 70, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Kohler, B.; Mueller-Roeber, B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010, 62, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.I.; Asensi-Fabado, M.A.; Munne-Bosch, S.; Antonio, C.; Tohge, T.; et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 2012, 24, 482–506. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.D.; Seo, P.J.; Yoon, H.K.; Park, C.M. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 2011, 23, 2155–2168. [Google Scholar] [CrossRef]
- Horsch, R.B.; Fry, J.E.; Hoffmann, D.; Eichholtz, D.; Rogers, S.G.; Fraley, R.T. A simple and general method for transferring genes into plants. Science 1985, 227, 1229–1231. [Google Scholar]
- Sierro, N.; Battey, J.N.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willig, A.; Goepfert, S.; Peitsch, M.C.; Ivanov, N.V. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Osaki, M.; Takebe, M.; Shinano, T.; Wasaki, J. Endogenous hormones and expression of senescence-related genes in different senescent types of maize. J. Exp. Bot. 2005, 56, 1117–1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, P.O.; Nam, H.G. The molecular and genetic control of leaf senescence and longevity in Arabidopsis. Curr. Top Dev. Biol. 2005, 67, 49–83. [Google Scholar] [PubMed]
- Guo, P.; Li, Z.; Huang, P.; Li, B.; Fang, S.; Chu, J.; Guo, H. A Tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 2017, 29, 2854–2870. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, C.; Wang, R.; Chen, Y.; Shu, S.; Huang, R.; Zhang, D.; Li, J.; Xiao, S.; Yao, N.; et al. The Arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-Dependent salicylic acid accumulation and signaling. Plant Physiol. 2017, 173, 2294–2307. [Google Scholar] [CrossRef]
- Zhang, K.; Xia, X.; Zhang, Y.; Gan, S.S. An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J. 2012, 69, 667–678. [Google Scholar] [CrossRef]
- Woo, H.R.; Kim, J.H.; Kim, J.; Lee, U.; Song, I.J.; Lee, H.Y.; Nam, H.G.; Lim, P.O. The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J. Exp. Bot. 2010, 61, 3947–3957. [Google Scholar] [CrossRef]
- Ghanem, M.E.; Albacete, A.; Martinez-Andujar, C.; Acosta, M.; Romero-Aranda, R.; Dodd, I.C.; Lutts, S.; Perez-Alfocea, F. Hormonal changes during salinity-induced leaf senescence in tomato (Solanum lycopersicum L.). J. Exp. Bot. 2008, 59, 3039–3050. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Joseph, L.M.; Deng, W.T.; Liu, L.; Li, Q.B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Y.; Tureckova, V.; Xue, G.P.; Fernie, A.R.; Mueller-Roeber, B.; Balazadeh, S. The NAC transcription factor SlNAP2 regulates leaf senescence and fruit yield in tomato. Plant Physiol. 2018, 177, 1286–1302. [Google Scholar] [CrossRef]
- Gonzalez-Guzman, M.; Apostolova, N.; Belles, J.M.; Barrero, J.M.; Piqueras, P.; Ponce, M.R.; Micol, J.L.; Serrano, R.; Rodriguez, P.L. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 2002, 14, 1833–1846. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.C.; Hwang, S.G.; Endo, A.; Okamoto, M.; Koshiba, T.; Cheng, W.H. Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiol. 2007, 143, 745–758. [Google Scholar] [CrossRef]
- North, H.M.; De Almeida, A.; Boutin, J.P.; Frey, A.; To, A.; Botran, L.; Sotta, B.; Marion-Poll, A. The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J. 2007, 50, 810–824. [Google Scholar] [CrossRef]
- Leung, J.; Merlot, S.; Giraudat, J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 1997, 9, 759–771. [Google Scholar] [CrossRef]
- Xiao, H.; Nassuth, A. Stress- and development-induced expression of spliced and unspliced transcripts from two highly similar dehydrin 1 genes in V. riparia and V. vinifera. Plant Cell Rep. 2006, 25, 968–977. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Huang, R. Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol. Biol. 2010, 73, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Reid, K.E.; Olsson, N.; Schlosser, J.; Peng, F.; Lund, S.T. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006, 6, 27. [Google Scholar] [CrossRef]
- Gutha, L.R.; Casassa, L.F.; Harbertson, J.F.; Naidu, R.A. Modulation of flavonoid biosynthetic pathway genes and anthocyanins due to virus infection in grapevine (Vitis vinifera L.) leaves. BMC Plant Biol. 2010, 10, 187. [Google Scholar] [CrossRef]
- Costa, M.D.; Reis, P.A.; Valente, M.A.; Irsigler, A.S.; Carvalho, C.M.; Loureiro, M.E.; Aragao, F.J.; Boston, R.S.; Fietto, L.G.; Fontes, E.P. A new branch of endoplasmic reticulum stress signaling and the osmotic signal converge on plant-specific asparagine-rich proteins to promote cell death. J. Biol. Chem. 2008, 283, 20209–20219. [Google Scholar] [CrossRef]
- Jiang, G.; Wang, Z.; Shang, H.; Yang, W.; Hu, Z.; Phillips, J.; Deng, X. Proteome analysis of leaves from the resurrection plant Boea hygrometrica in response to dehydration and rehydration. Planta 2007, 225, 1405–1420. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Gen. Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Pervaiz, T.; Haifeng, J.; Salman Haider, M.; Cheng, Z.; Cui, M.; Wang, M.; Cui, L.; Wang, X.; Fang, J. Transcriptomic analysis of grapevine (cv. Summer Black) leaf, using the illumina platform. PloS ONE 2016, 11, e0147369. [Google Scholar] [CrossRef]

| Gene Name | Forward Primer 5′→3′ | Reverse Primer 5′→3′ |
|---|---|---|
| NtNCED1 (NM_001325669) | gtttggtggggagcctctgttttta | tgatttccattctttctcattgtgc |
| NtNCED2 (NM_001326185) | tccacttcaaaaccaaccactatt | ttaaggcactttccacggcatcta |
| NtNCED3 (JX101473) | ctcacacgaactccaaaacccactt | tagcaccattacggacataaacccc |
| NtNCED5 (XM_016625112) | ctccaaaacccacttcctaaaaca | agcaccattacggacataaacccc |
| NtZEP1 (XM_016582459) | tagaggaccaattcagatacagagca | agttaccagaaacaccatcaaccaag |
| NtABA2 (EU123520) | ggtgctgatggcataaggtctaa | ctccacccacatctgaagaaaca |
| NtABA4 (XM_016637325) | cttccacttgcttttgtcactccc | cacccatttctatgcttacttccc |
| NtABI2 (NM_001324946) | tgttatgcagggtggtgttaaaggc | aaggtgaaggtgaaagagatgatgt |
| NtPYL4 (XP_00977852) | atcggtctgttactactttacatcct | cctttcgtctacccaaattctca |
| NtActin (XM_016618073) | tggagaaaatctggcatcacacg | actggcataaagggatagaacgg |
| VvDRL1 (XP-002281816) | ccgggtttcggtttcatccta | actccctctcgccaatcttcg |
| VvActin (AF369524) | cctcaaccccaaggccaacaga | accatcaccagaatccagcaca |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Z.; Li, G.; Yan, C.; Liu, L.; Zhang, Q.; Han, Z.; Li, B. DRL1, Encoding A NAC Transcription Factor, Is Involved in Leaf Senescence in Grapevine. Int. J. Mol. Sci. 2019, 20, 2678. https://doi.org/10.3390/ijms20112678
Zhu Z, Li G, Yan C, Liu L, Zhang Q, Han Z, Li B. DRL1, Encoding A NAC Transcription Factor, Is Involved in Leaf Senescence in Grapevine. International Journal of Molecular Sciences. 2019; 20(11):2678. https://doi.org/10.3390/ijms20112678
Chicago/Turabian StyleZhu, Ziguo, Guirong Li, Chaohui Yan, Li Liu, Qingtian Zhang, Zhen Han, and Bo Li. 2019. "DRL1, Encoding A NAC Transcription Factor, Is Involved in Leaf Senescence in Grapevine" International Journal of Molecular Sciences 20, no. 11: 2678. https://doi.org/10.3390/ijms20112678




