Development and Optimization of Expression, Purification, and ATPase Assay of KaiC for Medium-Throughput Screening of Circadian Clock Mutants in Cyanobacteria
Abstract
:1. Introduction
2. Results
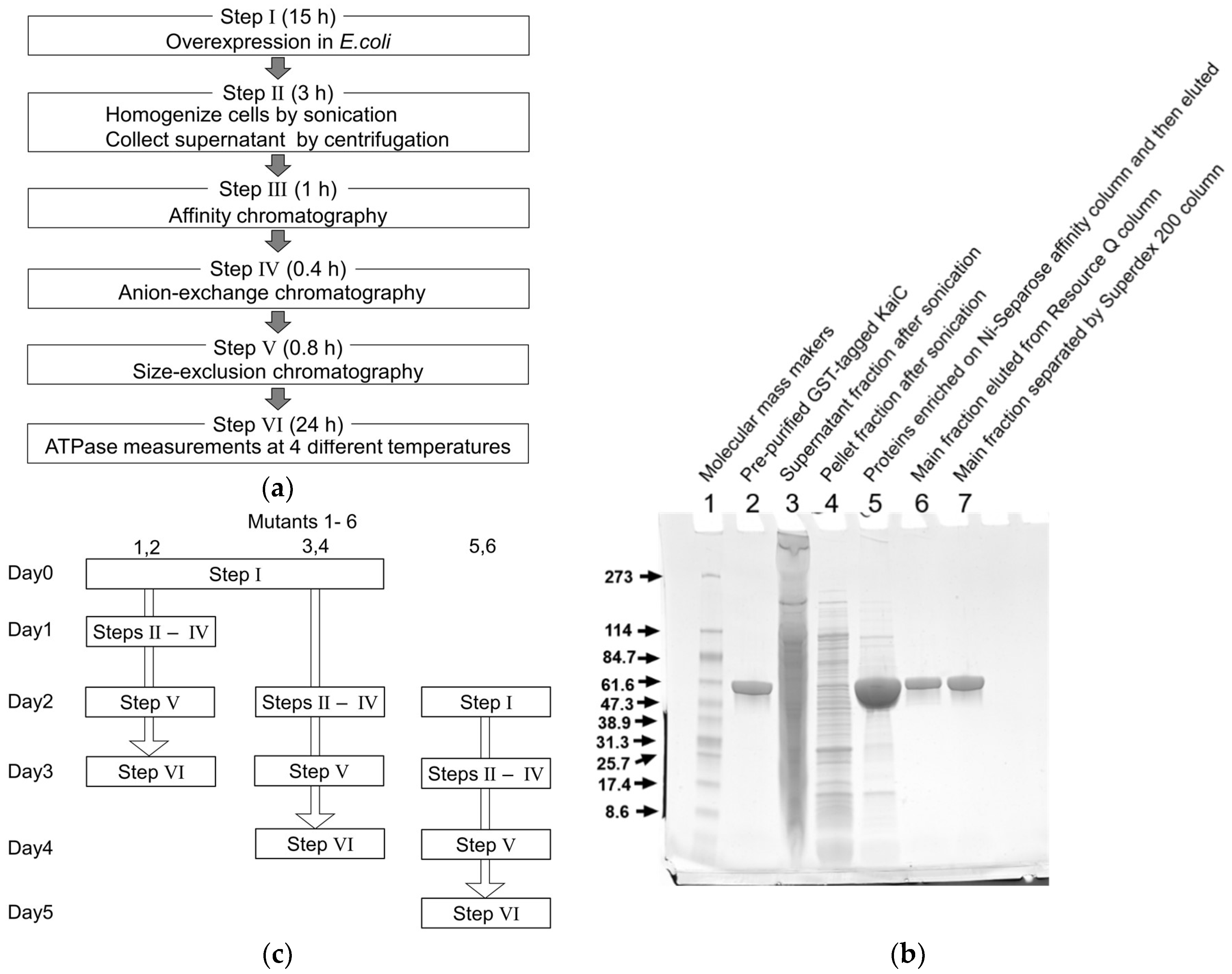
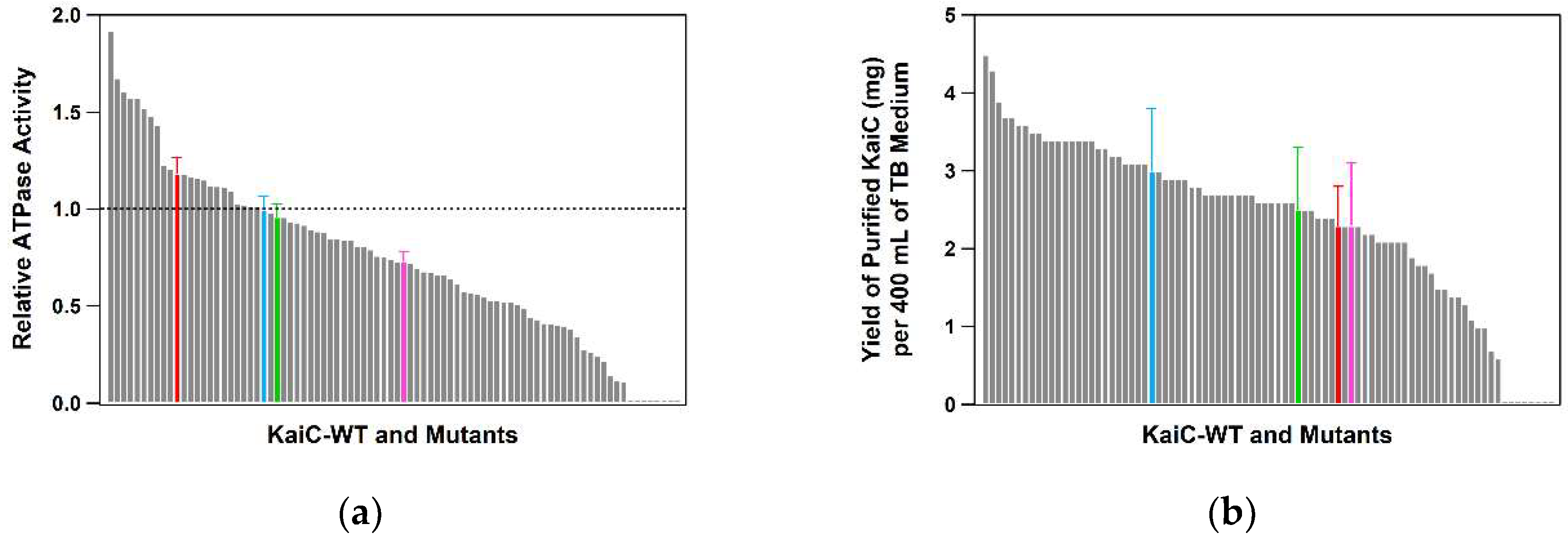
2.1. Optimization of the Expression and Purification of KaiC
2.2. Development of the HPLC System Equipped with a Multi-Channel High-Precision Temperature Controller
2.3. Throughput, Performance, and Reproducibility of the Protocol
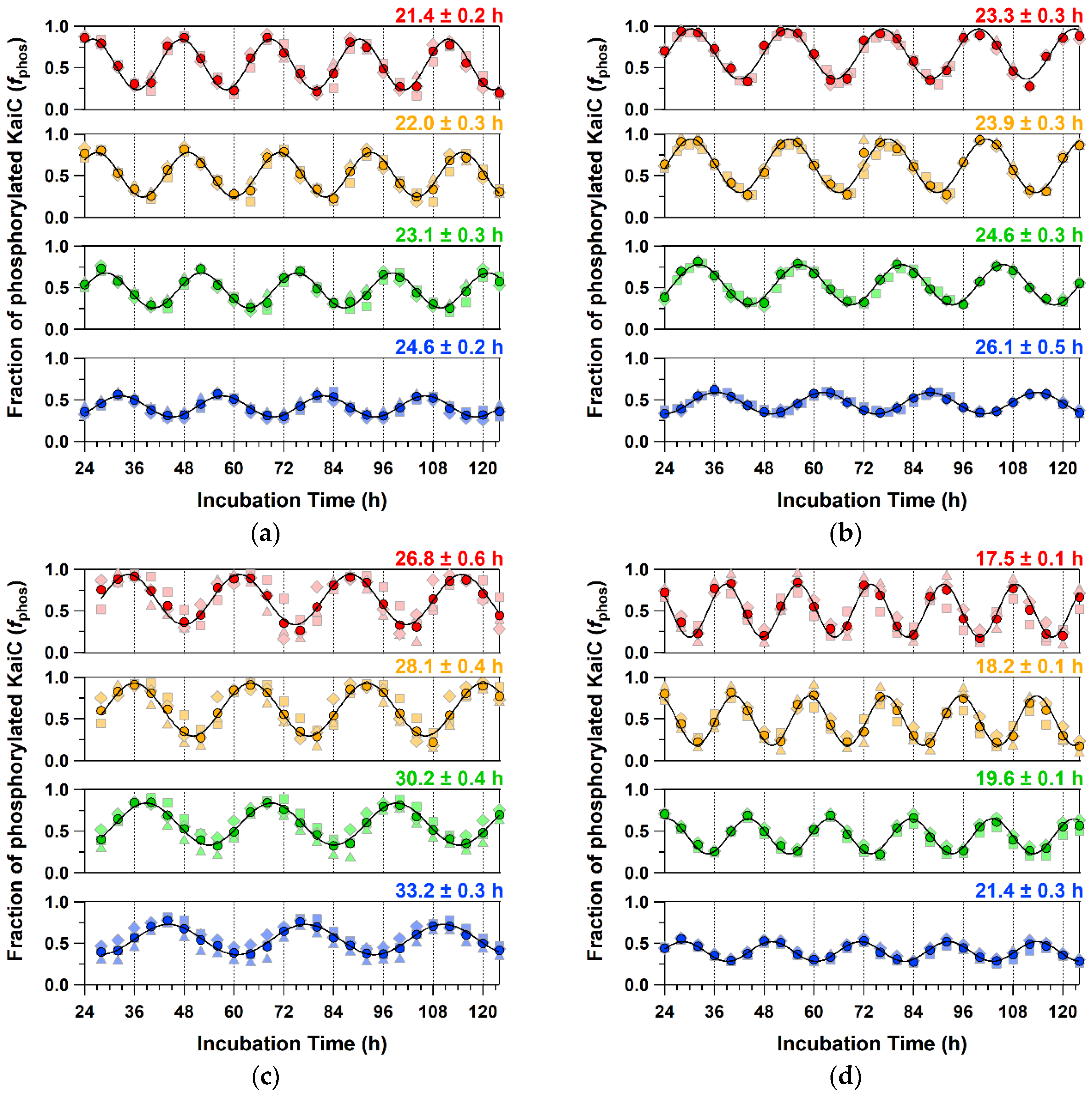
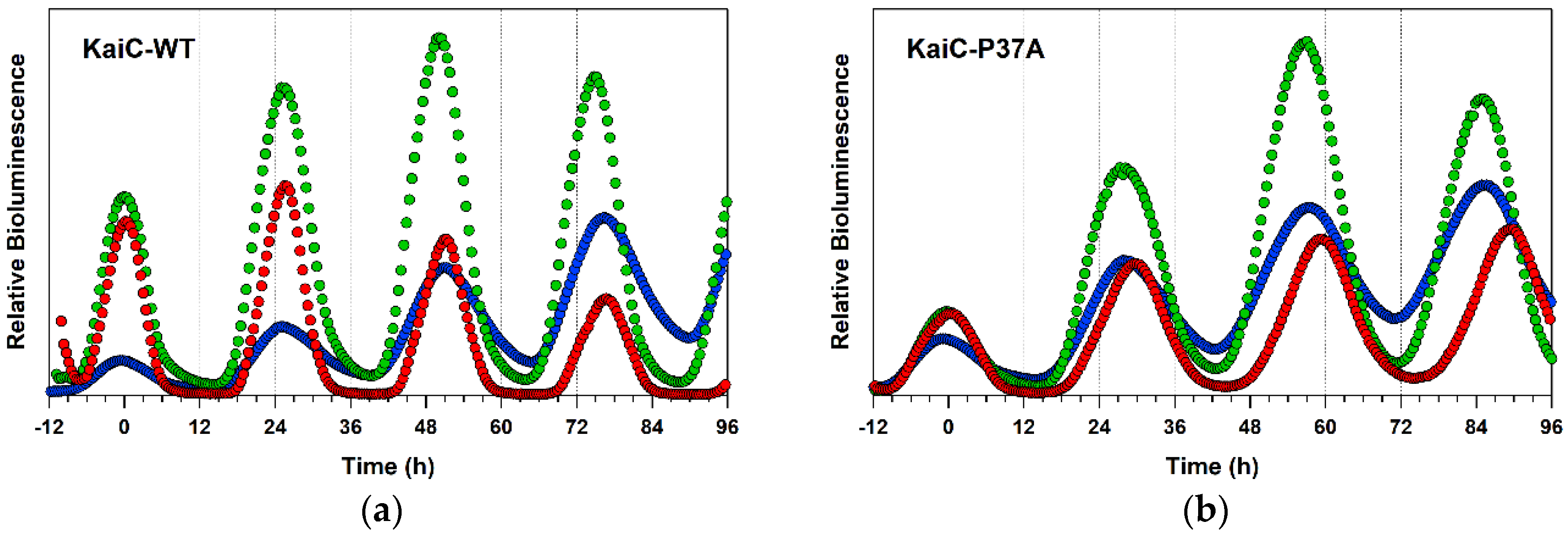
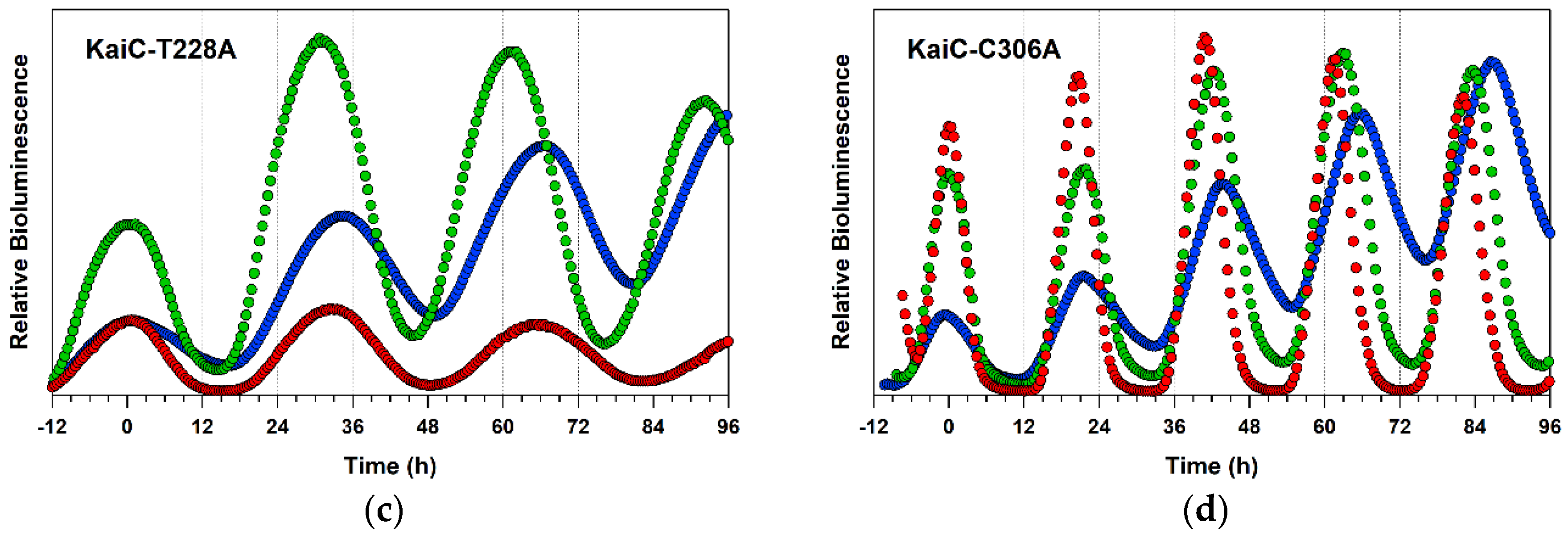
2.4. Detailed Characterizations of Representative ATPase Mutants of KaiC
3. Discussion
4. Materials and Methods
4.1. Expressions and Purifications
4.2. Development of Temperature-Controlled Sample Tables
4.3. ATPase Measurements
4.4. KaiC Phosphorylation/Dephosphorylation Cycle
4.5. In Vivo Bioluminsescence Assay
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HPLC | high-performance liquid chromatography |
| ATP | adenosine triphosphate |
| ADP | adenosine diphosphate |
| PTO | post-translational oscillation |
| TTO | transcriptional and translational oscillation |
References
- Pittendrigh, C.S. Temporal organization: Reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 1993, 55, 16–54. [Google Scholar] [CrossRef] [PubMed]
- Reppert, S.M.; Weaver, D.R. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 2001, 63, 647–676. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Golden, S.S.; Ishiura, M.; Kondo, T. Circadian clocks in prokaryotes. Mol. Microbiol. 1996, 21, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Elliott, J.; Foster, R.; Honma, K.; Kronauer, R. Fundamental Properties of Circadian Rhythms. In Chronobiology: Biological Timekeeping; Dunlap, J.C., Loros, J.J., DeCoursey, P.J., Eds.; Sinauer Associates, Inc. Publishers: Sunderland, MA, USA, 2004; Volume 3, pp. 67–105. ISBN -13. [Google Scholar]
- Young, M.W.; Kay, S.A. Time zones: A comparative genetics of circadian clocks. Nat. Rev. Genet. 2001, 2, 702–715. [Google Scholar] [CrossRef]
- O’Neill, J.S.; van Ooijen, G.; Dixon, L.E.; Troein, C.; Corellou, F.; Bouget, F.Y.; Reddy, A.B.; Millar, A.J. Circadian rhythms persist without transcription in a eukaryote. Nature 2011, 469, 554–558. [Google Scholar] [CrossRef] [PubMed]
- Ishiura, M.; Kutsuna, S.; Aoki, S.; Iwasaki, H.; Andersson, C.R.; Tanabe, A.; Golden, S.S.; Johnson, C.H.; Kondo, T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 1998, 281, 1519–1523. [Google Scholar] [CrossRef]
- Tomita, J.; Nakajima, M.; Kondo, T.; Iwasaki, H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 2005, 307, 251–254. [Google Scholar] [CrossRef]
- Nakajima, M.; Imai, K.; Ito, H.; Nishiwaki, T.; Murayama, Y.; Iwasaki, H.; Oyama, T.; Kondo, T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 2005, 308, 414–415. [Google Scholar] [CrossRef]
- Terauchi, K.; Kitayama, Y.; Nishiwaki, T.; Miwa, K.; Murayama, Y.; Oyama, T.; Kondo, T. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc. Natl. Acad. Sci USA 2007, 104, 16377–16381. [Google Scholar] [CrossRef] [Green Version]
- Kageyama, H.; Nishiwaki, T.; Nakajima, M.; Iwasaki, H.; Oyama, T.; Kondo, T. Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol. Cell 2006, 23, 161–171. [Google Scholar] [CrossRef]
- Akiyama, S.; Nohara, A.; Ito, K.; Maeda, Y. Assembly and disassembly dynamics of the cyanobacterial periodosome. Mol. Cell 2008, 29, 703–716. [Google Scholar] [CrossRef]
- Abe, J.; Hiyama, T.B.; Mukaiyama, A.; Son, S.; Mori, T.; Saito, S.; Osako, M.; Wolanin, J.; Yamashita, E.; Kondo, T.; et al. Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock. Science 2015, 349, 312–316. [Google Scholar] [CrossRef]
- Petegem, V.F.; Duderstadt, K.E.; Clark, K.A.; Wang, M.; Minor, D.L., Jr. Alanine-scanning mutagenesis defines a conserved energetic hotspot in the CaValpha1 AID-CaVbeta interaction site that is critical for channel modulation. Structure 2008, 16, 280–294. [Google Scholar] [CrossRef]
- Samai, P.; Shuman, S. Structure-function analysis of the OB and latch domains of chlorella virus DNA ligase. J. Biol. Chem. 2011, 286, 22642–22652. [Google Scholar] [CrossRef]
- Iwasaki, H.; Taniguchi, Y.; Ishiura, M.; Kondo, T. Physical interactions among circadian clock proteins KaiA, KaiB and KaiC in cyanobacteria. EMBO J. 1999, 18, 1137–1145. [Google Scholar] [CrossRef]
- Murayama, Y.; Mukaiyama, A.; Imai, K.; Onoue, Y.; Tsunoda, A.; Nohara, A.; Ishida, T.; Maeda, Y.; Terauchi, K.; Kondo, T.; et al. Tracking and visualizing the circadian ticking of the cyanobacterial clock protein KaiC in solution. EMBO J. 2011, 30, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Murayama, Y.; Kori, H.; Oshima, C.; Kondo, T.; Iwasaki, H.; Ito, H. Low temperature nullifies the circadian clock in cyanobacteria through Hopf bifurcation. Proc. Natl. Acad. Sci. USA 2017, 114, 5641–5646. [Google Scholar] [CrossRef] [Green Version]
- Kurosawa, G.; Fujioka, A.; Koinuma, S.; Mochizuki, A.; Shigeyoshi, Y. Temperature–amplitude coupling for stable biological rhythms at different temperatures. PLoS Comput. Biol. 2017, 13, e1005501. [Google Scholar] [CrossRef]
- Pattanayek, R.; Wang, J.; Mori, T.; Xu, Y.; Johnson, C.H.; Egli, M. Visualizing a circadian clock protein: Crystal structure of KaiC and functional insights. Mol. Cell 2004, 15, 375–388. [Google Scholar] [CrossRef]
- Nishiwaki, T.; Satomi, Y.; Nakajima, M.; Lee, C.; Kiyohara, R.; Kageyama, H.; Kitayama, Y.; Temamoto, M.; Yamaguchi, A.; Hijikata, A.; et al. Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc. Natl. Acad. Sci. USA 2004, 101, 13927–13932. [Google Scholar] [CrossRef]
- Furuike, Y.; Abe, J.; Mukaiyama, A.; Akiyama, S. Accelerating in vitro studies on circadian clock systems using an automated sampling device. Biophys. Phys. 2016, 13, 235–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiyama, S. Structural and dynamic aspects of protein clocks: How can they be so slow and stable? Cell Mol. Life Sci. 2012, 69, 2147–2160. [Google Scholar] [CrossRef] [PubMed]



| KaiC | In Vitro ATPase Activity 1,2 | In Vitro Phosphorylation Cycle 1,3 | In Vivo Bioluminescence Rhythm 1 | |||
|---|---|---|---|---|---|---|
| (d−1) 4 | Q10 | Period (h) 4 | Q10 | Period (h) 5 | Q106 | |
| WT | 13.4 ± 0.9 | 1.01 ± 0.04 | 23.1 ± 0.3 | 1.09 ± 0.00 | 24.8 ± 0.2 | 1.05 ± 0.01 (0.99 ± 0.01) |
| P37A | 12.9 ± 1.6 * | 1.03 ± 0.02 * | 24.6 ± 0.3 *** | 1.08 ± 0.00 *** | 28.1 ± 0.1 **** | 1.01 ± 0.01 *** (0.93 ± 0.01 ***) |
| T228A | 9.8 ± 0.9 *** | 1.18 ± 0.06 ** | 30.2 ± 0.4 **** | 1.15 ± 0.01 *** | 30.2 ± 0.2 **** | 1.17 ± 0.03 ** (0.94 ± 0.03 *) |
| C306A | 15.9 ± 2.2 * | 1.14 ± 0.05 ** | 19.6 ± 0.1 **** | 1.14 ± 0.02 ** | 20.8 ± 0.0 **** | 1.09 ± 0.01 *** (1.05 ± 0.01 ****) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ouyang, D.; Furuike, Y.; Mukaiyama, A.; Ito-Miwa, K.; Kondo, T.; Akiyama, S. Development and Optimization of Expression, Purification, and ATPase Assay of KaiC for Medium-Throughput Screening of Circadian Clock Mutants in Cyanobacteria. Int. J. Mol. Sci. 2019, 20, 2789. https://doi.org/10.3390/ijms20112789
Ouyang D, Furuike Y, Mukaiyama A, Ito-Miwa K, Kondo T, Akiyama S. Development and Optimization of Expression, Purification, and ATPase Assay of KaiC for Medium-Throughput Screening of Circadian Clock Mutants in Cyanobacteria. International Journal of Molecular Sciences. 2019; 20(11):2789. https://doi.org/10.3390/ijms20112789
Chicago/Turabian StyleOuyang, Dongyan, Yoshihiko Furuike, Atsushi Mukaiyama, Kumiko Ito-Miwa, Takao Kondo, and Shuji Akiyama. 2019. "Development and Optimization of Expression, Purification, and ATPase Assay of KaiC for Medium-Throughput Screening of Circadian Clock Mutants in Cyanobacteria" International Journal of Molecular Sciences 20, no. 11: 2789. https://doi.org/10.3390/ijms20112789





