Post-Translational Modifications of Proteins Have Versatile Roles in Regulating Plant Immune Responses
Abstract
:1. Introduction
2. Many Protein Effectors Function through PTMs of Host Proteins in Plant Cells
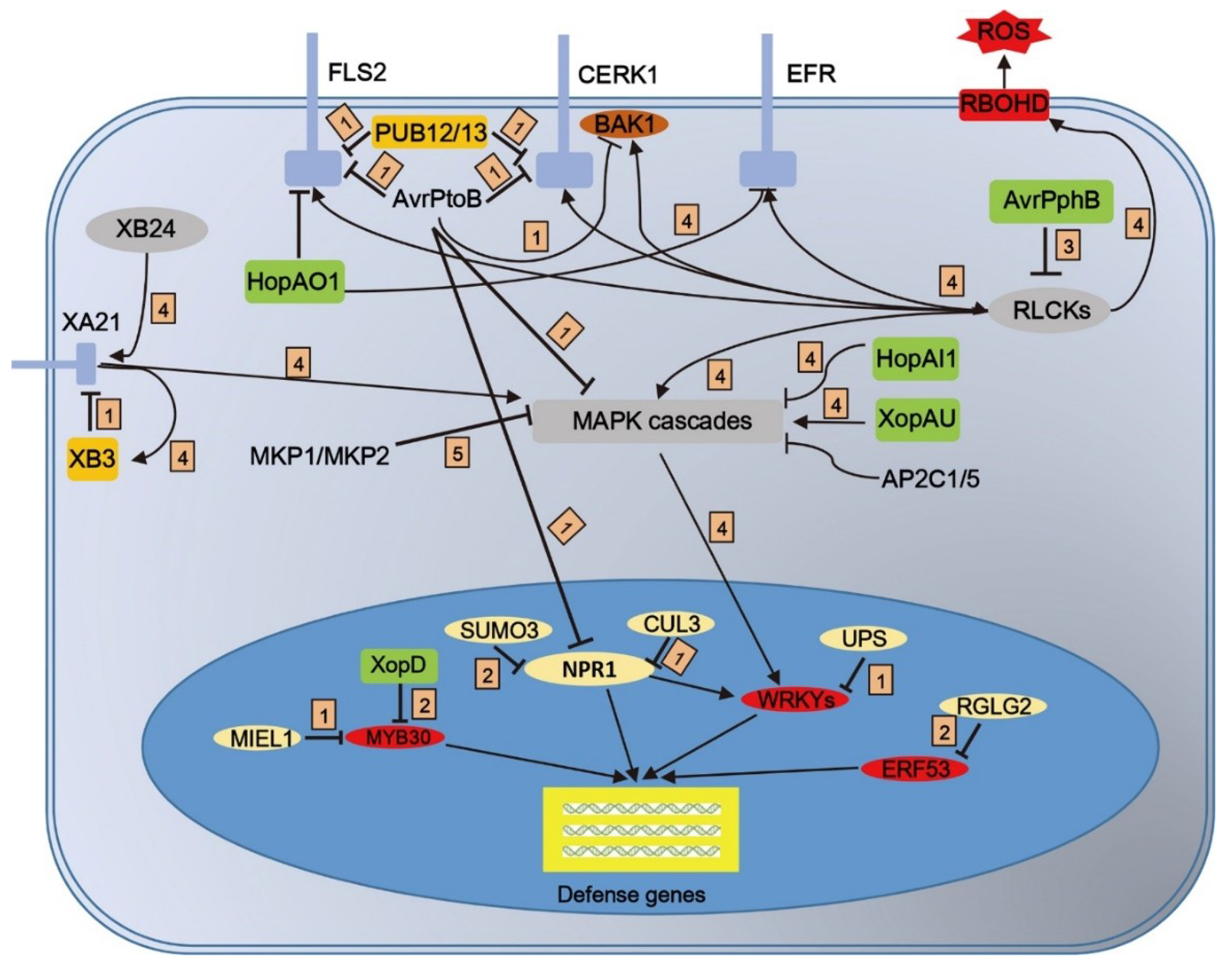
3. PTMs of Receptor Complexes Initiate Plant Immune Signaling
4. PTMs of RLCK–MAPK Are Required for Immune Signal Transduction
5. PTM Modulates TF Activities in Plant Immunity
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AP2C1 | Arabidopsis PP2C-type phosphatase |
| AVR | avirulence |
| BAK1 | BRI1-associated receptor kinase1 |
| BIK1 | botrytis-induced kinase1 |
| BR | brassinosteroid |
| BTB/POZ | broad-complex, tramtrack, and bric-à-brac/poxvirus, zinc finger |
| bZIP | basic leucine zipper |
| CEBiP | chitin elicitor-binding protein |
| CERK1 | chitin elicitor receptor kinase 1 |
| ECD | extracellular domain |
| EFR | EF-TU receptor |
| ETI | effector-triggered immunity |
| ERF | ET responsive TF |
| ER-QC | endoplasmic reticulum-quality control |
| FLS2 | Flagellin Sensing2 |
| HR | hypersensitive response |
| IPA1 | ideal plant architecture 1 |
| LRR | leucine-rich repeat |
| LYK5 | lysine motif receptor kinase 5 |
| LYP4 | lysin motif-containing protein 4 |
| MAMPs | microbe associated molecular patterns |
| MAPK | mitogen-activated protein kinase |
| MAPKK/MKK/MEK | MAPK kinase |
| MAPKKK/MEKK | MAPK kinase kinase |
| MIEL1 | MYB30-interacting E3 ligase1 |
| MKS1 | MAPK4 substrate 1 |
| MYB | myeloblastosis |
| NAC | no apical meristem, Arabidopsis transcription activation factor, cup-shaped cotyledon |
| NLRs | NOD-like receptors |
| NOD-like receptors | nucleotide-binding oligomerization domain-like receptors |
| P. syringae | Pseudomonas syringae |
| PAMPs | pathogen associated molecular patterns |
| PBL2 | PBS1-like protein 2 |
| PCD | programmed cell death |
| PCRK1 | PTI compromised receptor-like cytoplasmic kinase1 |
| PGN | eptidoglycan |
| PP2C | protein phosphatase 2C |
| PRRs | PAMPs are detected by pattern-recognition receptors |
| PTI | PAMP-/MAMP-triggered immunity |
| PTMs | post-translational modifications |
| PUB | plant U-box |
| RBOHD | respiratory burst oxidase homolog protein D |
| R protein | resistance protein |
| RLCKs | receptor-like cytoplasmic kinases |
| RIN4 | RPM1-interacting protein 4 |
| RLKs | receptor-like kinases |
| RLPs | receptor-like pro2teins |
| ROS | reactive oxygen species |
| RKS1 | resistance-related kinase 1 |
| SAR | systemic acquired resistance |
| Ser | serine |
| SERKs | somatic embryogenesis receptor kinases |
| SDS2 | SPL11 cell-death suppressor 2 |
| SIPK | SA-induced protein kinase |
| SPL | SQUAMOSA promoter-binding protein-like |
| SUMO | small Ub-like modifier |
| Thr | threonine |
| Tyr | tyrosine |
| XB3 | XA21-Bingding Protein3 |
| Xoo | Xanthomonas oryzae pv. Oryzae |
| ZAR1 | HOPZ-activated resistance 1 |
References
- Wei, T.; Chern, M.; Liu, F.; Ronald, P.C. Suppression of bacterial infection in rice by treatment with a sulfated peptide. Mol. Plant Pathol. 2016, 17, 1493–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Boutrot, F.; Zipfel, C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Ann. Rev. Phytopathol. 2017, 55, 257–286. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.W.; Klessig, D.F. Damps, mamps, and namps in plant innate immunity. BMC Plant Biol. 2016, 16, 232. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yu, Y.; Zhou, Z.; Zhou, J.M. Plant pattern-recognition receptors controlling innate immunity. Sci. China Life Sci. 2016, 59, 878–888. [Google Scholar] [CrossRef] [Green Version]
- Gust, A.A.; Felix, G. Receptor like proteins associate with sobir1-type of adaptors to form bimolecular receptor kinases. Curr. Opin. Plant Biol. 2014, 21, 104–111. [Google Scholar] [CrossRef]
- Liebrand, T.W.; van den Burg, H.A.; Joosten, M.H. Two for all: Receptor-associated kinases sobir1 and bak1. Trends Plant Sci. 2014, 19, 123–132. [Google Scholar] [CrossRef]
- Macho, A.P.; Zipfel, C. Plant prrs and the activation of innate immune signaling. Mol. Cell 2014, 54, 263–272. [Google Scholar] [CrossRef]
- Gomez-Gomez, L.; Boller, T. Fls2: An lrr receptor-like kinase involved in the perception of the bacterial elicitor flagellin in arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Chinchilla, D.; Boller, T.; Robatzek, S. Flagellin signalling in plant immunity. Adv. Exp. Med. Biol. 2007, 598, 358–371. [Google Scholar]
- Tsuda, K.; Sato, M.; Glazebrook, J.; Cohen, J.D.; Katagiri, F. Interplay between mamp-triggered and sa-mediated defense responses. Plant J. Cell Mol. Biol. 2008, 53, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Fiorin, G.L.; Sanchez-Vallet, A.; Thomazella, D.P.T.; do Prado, P.F.V.; do Nascimento, L.C.; Figueira, A.V.O.; Thomma, B.; Pereira, G.A.G.; Teixeira, P. Suppression of plant immunity by fungal chitinase-like effectors. Curr. Biol. CB 2018, 28, 3023–3030.e3025. [Google Scholar] [CrossRef] [PubMed]
- Houterman, P.M.; Cornelissen, B.J.; Rep, M. Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog. 2008, 4, e1000061. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacob, F.; Vernaldi, S.; Maekawa, T. Evolution and conservation of plant nlr functions. Front. Immunol. 2013, 4, 297. [Google Scholar] [CrossRef] [PubMed]
- Coll, N.S.; Epple, P.; Dangl, J.L. Programmed cell death in the plant immune system. Cell Death Differ. 2011, 18, 1247–1256. [Google Scholar] [CrossRef] [Green Version]
- Mur, L.A.; Kenton, P.; Lloyd, A.J.; Ougham, H.; Prats, E. The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 2008, 59, 501–520. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, T.; Kufer, T.A.; Schulze-Lefert, P. Nlr functions in plant and animal immune systems: So far and yet so close. Nat. Immunol. 2011, 12, 817–826. [Google Scholar] [CrossRef]
- Griebel, T.; Maekawa, T.; Parker, J.E. Nod-like receptor cooperativity in effector-triggered immunity. Trends Immunol. 2014, 35, 562–570. [Google Scholar] [CrossRef]
- Withers, J.; Dong, X. Post-translational regulation of plant immunity. Curr. Opin. Plant Biol. 2017, 38, 124–132. [Google Scholar] [CrossRef]
- Stulemeijer, I.J.; Joosten, M.H. Post-translational modification of host proteins in pathogen-triggered defence signalling in plants. Mol. Plant Pathol. 2008, 9, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Yang, W. E3 ubiquitin ligases: Ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 2017, 58, 1461–1476. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Joshi, D.; Yadav, P.K.; Gupta, A.K.; Bhatt, T.K. Role of ubiquitin-mediated degradation system in plant biology. Front. Plant Sci. 2016, 7, 806. [Google Scholar] [CrossRef] [PubMed]
- Van den Burg, H.A.; Kini, R.K.; Schuurink, R.C.; Takken, F.L. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell 2010, 22, 1998–2016. [Google Scholar] [CrossRef] [PubMed]
- Augustine, R.C.; Vierstra, R.D. Sumoylation: Re-wiring the plant nucleus during stress and development. Curr. Opin. Plant Biol. 2018, 45, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Grzonka, Z.; Kasprzykowski, F.; Wiczk, W. Cysteine proteases. In Industrial Enzymes: Structure, Function and Applications; Polaina, J., MacCabe, A.P., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2007; pp. 181–195. [Google Scholar]
- Roth, J.; Zuber, C.; Park, S.; Jang, I.; Lee, Y.; Kysela, K.G.; Le Fourn, V.; Santimaria, R.; Guhl, B.; Cho, J.W. Protein n-glycosylation, protein folding, and protein quality control. Mol. Cells 2010, 30, 497–506. [Google Scholar] [CrossRef]
- Uhse, S.; Djamei, A. Effectors of plant-colonizing fungi and beyond. PLoS Pathog. 2018, 14, e1006992. [Google Scholar] [CrossRef]
- Collemare, J.; O’Connell, R.; Lebrun, M.H. Nonproteinaceous effectors: The terra incognita of plant-fungal interactions. New Phytol. 2019. [Google Scholar] [CrossRef]
- Lievens, L.; Pollier, J.; Goossens, A.; Beyaert, R.; Staal, J. Abscisic acid as pathogen effector and immune regulator. Front. Plant Sci. 2017, 8, 587. [Google Scholar] [CrossRef]
- Audenaert, K.; Vanheule, A.; Hofte, M.; Haesaert, G. Deoxynivalenol: A major player in the multifaceted response of fusarium to its environment. Toxins 2013, 6, 1–19. [Google Scholar] [CrossRef]
- Wight, W.D.; Kim, K.H.; Lawrence, C.B.; Walton, J.D. Biosynthesis and role in virulence of the histone deacetylase inhibitor depudecin from alternaria brassicicola. Mol. Plant Microbe Interact. 2009, 22, 1258–1267. [Google Scholar] [CrossRef] [PubMed]
- Wicklow, D.T.; Jordan, A.M.; Gloer, J.B. Antifungal metabolites (monorden, monocillins i, ii, iii) from colletotrichum graminicola, a systemic vascular pathogen of maize. Mycol. Res. 2009, 113, 1433–1442. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Huang, Y.; Fang, M.; Zhang, Y.; Zheng, Z.; Zhao, Y.; Su, W. Brefeldin a, a cytotoxin produced by paecilomyces sp. And aspergillus clavatus isolated from taxus mairei and torreya grandis. FEMS Immunol. Med. Microbiol. 2002, 34, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Kusch, S.; Frantzeskakis, L.; Thieron, H.; Panstruga, R. Small rnas from cereal powdery mildew pathogens may target host plant genes. Fungal Biol. 2018, 122, 1050–1063. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Weiberg, A.; Dellota, E., Jr.; Yamane, D.; Jin, H. Botrytis small rna bc-sir37 suppresses plant defense genes by cross-kingdom rnai. RNA Biol. 2017, 14, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Weiberg, A.; Wang, M.; Lin, F.M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.D.; Jin, H. Fungal small rnas suppress plant immunity by hijacking host rna interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Carella, P.; Evangelisti, E.; Schornack, S. Sticking to it: Phytopathogen effector molecules may converge on evolutionarily conserved host targets in green plants. Curr. Opin. Plant Biol. 2018, 44, 175–180. [Google Scholar] [CrossRef]
- Gohre, V.; Spallek, T.; Haweker, H.; Mersmann, S.; Mentzel, T.; Boller, T.; de Torres, M.; Mansfield, J.W.; Robatzek, S. Plant pattern-recognition receptor fls2 is directed for degradation by the bacterial ubiquitin ligase avrptob. Curr. Biol. CB 2008, 18, 1824–1832. [Google Scholar] [CrossRef]
- Shan, L.; He, P.; Li, J.; Heese, A.; Peck, S.C.; Nurnberger, T.; Martin, G.B.; Sheen, J. Bacterial effectors target the common signaling partner bak1 to disrupt multiple mamp receptor-signaling complexes and impede plant immunity. Cell Host Microbe 2008, 4, 17–27. [Google Scholar] [CrossRef]
- Gimenez-Ibanez, S.; Hann, D.R.; Ntoukakis, V.; Petutschnig, E.; Lipka, V.; Rathjen, J.P. Avrptob targets the lysm receptor kinase cerk1 to promote bacterial virulence on plants. Curr. Biol. CB 2009, 19, 423–429. [Google Scholar] [CrossRef]
- Zeng, L.; Velasquez, A.C.; Munkvold, K.R.; Zhang, J.; Martin, G.B. A tomato lysm receptor-like kinase promotes immunity and its kinase activity is inhibited by avrptob. Plant J. Cell Mol. Biol. 2012, 69, 92–103. [Google Scholar] [CrossRef]
- Mathieu, J.; Schwizer, S.; Martin, G.B. Pto kinase binds two domains of avrptob and its proximity to the effector e3 ligase determines if it evades degradation and activates plant immunity. PLoS Pathog. 2014, 10, e1004227. [Google Scholar] [CrossRef]
- Chen, H.; Chen, J.; Li, M.; Chang, M.; Xu, K.; Shang, Z.; Zhao, Y.; Palmer, I.; Zhang, Y.; McGill, J.; et al. A bacterial type iii effector targets the master regulator of salicylic acid signaling, npr1, to subvert plant immunity. Cell Host Microbe 2017, 22, 777–788.e7. [Google Scholar] [CrossRef]
- Wei, H.L.; Chakravarthy, S.; Mathieu, J.; Helmann, T.C.; Stodghill, P.; Swingle, B.; Martin, G.B.; Collmer, A. Pseudomonas syringae pv. Tomato dc3000 type iii secretion effector polymutants reveal an interplay between hopad1 and avrptob. Cell Host Microbe 2015, 17, 752–762. [Google Scholar] [CrossRef]
- Popov, G.; Majhi, B.B.; Sessa, G. Effector gene xopae of xanthomonas euvesicatoria 85-10 is part of an operon and encodes an e3 ubiquitin ligase. J. Bacteriol. 2018, 200, e00104-18. [Google Scholar] [CrossRef]
- Qin, J.; Zhou, X.; Sun, L.; Wang, K.; Yang, F.; Liao, H.; Rong, W.; Yin, J.; Chen, H.; Chen, X.; et al. The xanthomonas effector xopk harbours e3 ubiquitin-ligase activity that is required for virulence. New Phytol. 2018, 220, 219–231. [Google Scholar] [CrossRef]
- Kim, J.G.; Stork, W.; Mudgett, M.B. Xanthomonas type iii effector xopd desumoylates tomato transcription factor slerf4 to suppress ethylene responses and promote pathogen growth. Cell Host Microbe 2013, 13, 143–154. [Google Scholar] [CrossRef]
- Kim, J.G.; Taylor, K.W.; Hotson, A.; Keegan, M.; Schmelz, E.A.; Mudgett, M.B. Xopd sumo protease affects host transcription, promotes pathogen growth, and delays symptom development in xanthomonas-infected tomato leaves. Plant Cell 2008, 20, 1915–1929. [Google Scholar] [CrossRef]
- Canonne, J.; Marino, D.; Jauneau, A.; Pouzet, C.; Briere, C.; Roby, D.; Rivas, S. The xanthomonas type iii effector xopd targets the arabidopsis transcription factor myb30 to suppress plant defense. Plant Cell 2011, 23, 3498–3511. [Google Scholar] [CrossRef]
- Axtell, M.J.; Staskawicz, B.J. Initiation of rps2-specified disease resistance in arabidopsis is coupled to the avrrpt2-directed elimination of rin4. Cell 2003, 112, 369–377. [Google Scholar] [CrossRef]
- Afzal, A.J.; da Cunha, L.; Mackey, D. Separable fragments and membrane tethering of arabidopsis rin4 regulate its suppression of pamp-triggered immunity. Plant Cell 2011, 23, 3798–3811. [Google Scholar] [CrossRef]
- Coaker, G.; Falick, A.; Staskawicz, B. Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 2005, 308, 548–550. [Google Scholar] [CrossRef]
- Eschen-Lippold, L.; Jiang, X.; Elmore, J.M.; Mackey, D.; Shan, L.; Coaker, G.; Scheel, D.; Lee, J. Bacterial avrrpt2-like cysteine proteases block activation of the arabidopsis mitogen-activated protein kinases, mpk4 and mpk11. Plant Physiol. 2016, 171, 2223–2238. [Google Scholar] [CrossRef]
- Liang, X.; Zhou, J.M. Receptor-like cytoplasmic kinases: Central players in plant receptor kinase-mediated signaling. Ann. Rev. Plant Biol. 2018, 69, 267–299. [Google Scholar] [CrossRef]
- Zhang, J.; Li, W.; Xiang, T.; Liu, Z.; Laluk, K.; Ding, X.; Zou, Y.; Gao, M.; Zhang, X.; Chen, S.; et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a pseudomonas syringae effector. Cell Host Microbe 2010, 7, 290–301. [Google Scholar] [CrossRef]
- Teper, D.; Girija, A.M.; Bosis, E.; Popov, G.; Savidor, A.; Sessa, G. The xanthomonas euvesicatoria type iii effector xopau is an active protein kinase that manipulates plant map kinase signaling. PLoS Pathog. 2018, 14, e1006880. [Google Scholar] [CrossRef]
- Zhang, J.; Shao, F.; Li, Y.; Cui, H.; Chen, L.; Li, H.; Zou, Y.; Long, C.; Lan, L.; Chai, J.; et al. A pseudomonas syringae effector inactivates mapks to suppress pamp-induced immunity in plants. Cell Host Microbe 2007, 1, 175–185. [Google Scholar] [CrossRef]
- Macho, A.P.; Schwessinger, B.; Ntoukakis, V.; Brutus, A.; Segonzac, C.; Roy, S.; Kadota, Y.; Oh, M.H.; Sklenar, J.; Derbyshire, P.; et al. A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science 2014, 343, 1509–1512. [Google Scholar] [CrossRef]
- Van de Wouw, A.P.; Cozijnsen, A.J.; Hane, J.K.; Brunner, P.C.; McDonald, B.A.; Oliver, R.P.; Howlett, B.J. Evolution of linked avirulence effectors in leptosphaeria maculans is affected by genomic environment and exposure to resistance genes in host plants. PLoS Pathog. 2010, 6, e1001180. [Google Scholar] [CrossRef]
- Zong, N.; Xiang, T.; Zou, Y.; Chai, J.; Zhou, J.M. Blocking and triggering of plant immunity by pseudomonas syringae effector avrpto. Plant Signal. Behav. 2008, 3, 583–585. [Google Scholar] [CrossRef]
- Feng, F.; Yang, F.; Rong, W.; Wu, X.; Zhang, J.; Chen, S.; He, C.; Zhou, J.M. A xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 2012, 485, 114–118. [Google Scholar] [CrossRef]
- Wang, G.; Roux, B.; Feng, F.; Guy, E.; Li, L.; Li, N.; Zhang, X.; Lautier, M.; Jardinaud, M.F.; Chabannes, M.; et al. The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe 2015, 18, 285–295. [Google Scholar] [CrossRef]
- Chinchilla, D.; Zipfel, C.; Robatzek, S.; Kemmerling, B.; Nurnberger, T.; Jones, J.D.; Felix, G.; Boller, T. A flagellin-induced complex of the receptor fls2 and bak1 initiates plant defence. Nature 2007, 448, 497–500. [Google Scholar] [CrossRef]
- Yasuda, S.; Okada, K.; Saijo, Y. A look at plant immunity through the window of the multitasking coreceptor bak1. Curr. Opin. Plant Biol. 2017, 38, 10–18. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Z.; Liu, D.; Xu, J.; Wei, X.; Yan, L.; Yang, C.; Lou, Z.; Shui, W. Assessment of bak1 activity in different plant receptor-like kinase complexes by quantitative profiling of phosphorylation patterns. J. Proteomics 2014, 108, 484–493. [Google Scholar] [CrossRef]
- Perraki, A.; DeFalco, T.A.; Derbyshire, P.; Avila, J.; Sere, D.; Sklenar, J.; Qi, X.; Stransfeld, L.; Schwessinger, B.; Kadota, Y.; et al. Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature 2018, 561, 248–252. [Google Scholar] [CrossRef]
- Lu, D.; Wu, S.; Gao, X.; Zhang, Y.; Shan, L.; He, P. A receptor-like cytoplasmic kinase, bik1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 496–501. [Google Scholar] [CrossRef]
- Kadota, Y.; Sklenar, J.; Derbyshire, P.; Stransfeld, L.; Asai, S.; Ntoukakis, V.; Jones, J.D.; Shirasu, K.; Menke, F.; Jones, A.; et al. Direct regulation of the nadph oxidase rbohd by the prr-associated kinase bik1 during plant immunity. Mol. Cell 2014, 54, 43–55. [Google Scholar] [CrossRef]
- Li, L.; Li, M.; Yu, L.; Zhou, Z.; Liang, X.; Liu, Z.; Cai, G.; Gao, L.; Zhang, X.; Wang, Y.; et al. The fls2-associated kinase bik1 directly phosphorylates the nadph oxidase rbohd to control plant immunity. Cell Host Microbe 2014, 15, 329–338. [Google Scholar] [CrossRef]
- Kong, Q.; Sun, T.; Qu, N.; Ma, J.; Li, M.; Cheng, Y.T.; Zhang, Q.; Wu, D.; Zhang, Z.; Zhang, Y. Two redundant receptor-like cytoplasmic kinases function downstream of pattern recognition receptors to regulate activation of sa biosynthesis. Plant Physiol. 2016, 171, 1344–1354. [Google Scholar] [CrossRef]
- Desaki, Y.; Miyata, K.; Suzuki, M.; Shibuya, N.; Kaku, H. Plant immunity and symbiosis signaling mediated by lysm receptors. Innate Immun. 2018, 24, 92–100. [Google Scholar] [CrossRef]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.T.; Jedrzejczak, R.P.; Joachimiak, A.; Stacey, G. The kinase lyk5 is a major chitin receptor in arabidopsis and forms a chitin-induced complex with related kinase cerk1. Elife 2014, 3, e03766. [Google Scholar] [CrossRef]
- Yamada, K.; Yamaguchi, K.; Shirakawa, T.; Nakagami, H.; Mine, A.; Ishikawa, K.; Fujiwara, M.; Narusaka, M.; Narusaka, Y.; Ichimura, K.; et al. The arabidopsis cerk1-associated kinase pbl27 connects chitin perception to mapk activation. EMBO J. 2016, 35, 2468–2483. [Google Scholar] [CrossRef]
- Kaku, H.; Nishizawa, Y.; Ishii-Minami, N.; Akimoto-Tomiyama, C.; Dohmae, N.; Takio, K.; Minami, E.; Shibuya, N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 11086–11091. [Google Scholar] [CrossRef] [Green Version]
- Fan, J.; Bai, P.; Ning, Y.; Wang, J.; Shi, X.; Xiong, Y.; Zhang, K.; He, F.; Zhang, C.; Wang, R.; et al. The monocot-specific receptor-like kinase sds2 controls cell death and immunity in rice. Cell Host Microbe 2018, 23, 498–510.e5. [Google Scholar] [CrossRef]
- Lu, D.; Lin, W.; Gao, X.; Wu, S.; Cheng, C.; Avila, J.; Heese, A.; Devarenne, T.P.; He, P.; Shan, L. Direct ubiquitination of pattern recognition receptor fls2 attenuates plant innate immunity. Science 2011, 332, 1439–1442. [Google Scholar] [CrossRef]
- Liao, D.; Cao, Y.; Sun, X.; Espinoza, C.; Nguyen, C.T.; Liang, Y.; Stacey, G. Arabidopsis e3 ubiquitin ligase plant u-box13 (pub13) regulates chitin receptor lysin motif receptor kinase5 (lyk5) protein abundance. New Phytol. 2017, 214, 1646–1656. [Google Scholar] [CrossRef]
- Wang, Y.S.; Pi, L.Y.; Chen, X.; Chakrabarty, P.K.; Jiang, J.; De Leon, A.L.; Liu, G.Z.; Li, L.; Benny, U.; Oard, J.; et al. Rice xa21 binding protein 3 is a ubiquitin ligase required for full xa21-mediated disease resistance. Plant Cell 2006, 18, 3635–3646. [Google Scholar] [CrossRef]
- Nagashima, Y.; von Schaewen, A.; Koiwa, H. Function of n-glycosylation in plants. Plant Sci. 2018, 274, 70–79. [Google Scholar] [CrossRef]
- Haweker, H.; Rips, S.; Koiwa, H.; Salomon, S.; Saijo, Y.; Chinchilla, D.; Robatzek, S.; von Schaewen, A. Pattern recognition receptors require n-glycosylation to mediate plant immunity. J. Biol. Chem. 2010, 285, 4629–4636. [Google Scholar] [CrossRef]
- Li, J.; Zhao-Hui, C.; Batoux, M.; Nekrasov, V.; Roux, M.; Chinchilla, D.; Zipfel, C.; Jones, J.D. Specific er quality control components required for biogenesis of the plant innate immune receptor efr. Proc. Natl. Acad. Sci. USA 2009, 106, 15973–15978. [Google Scholar] [CrossRef]
- Nekrasov, V.; Li, J.; Batoux, M.; Roux, M.; Chu, Z.H.; Lacombe, S.; Rougon, A.; Bittel, P.; Kiss-Papp, M.; Chinchilla, D.; et al. Control of the pattern-recognition receptor efr by an er protein complex in plant immunity. EMBO J. 2009, 28, 3428–3438. [Google Scholar] [CrossRef]
- Saijo, Y.; Tintor, N.; Lu, X.; Rauf, P.; Pajerowska-Mukhtar, K.; Haweker, H.; Dong, X.; Robatzek, S.; Schulze-Lefert, P. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009, 28, 3439–3449. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.; Zhang, S. Mapk cascades in plant disease resistance signaling. Ann. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef]
- Zhang, M.; Su, J.; Zhang, Y.; Xu, J.; Zhang, S. Conveying endogenous and exogenous signals: Mapk cascades in plant growth and defense. Curr. Opin. Plant Biol. 2018, 45, 1–10. [Google Scholar] [CrossRef]
- Mizoguchi, T.; Ichimura, K.; Irie, K.; Morris, P.; Giraudat, J.; Matsumoto, K.; Shinozaki, K. Identification of a possible map kinase cascade in arabidopsis thaliana based on pairwise yeast two-hybrid analysis and functional complementation tests of yeast mutants. FEBS Lett. 1998, 437, 56–60. [Google Scholar] [CrossRef]
- Huang, Y.; Li, H.; Gupta, R.; Morris, P.C.; Luan, S.; Kieber, J.J. Atmpk4, an arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by atmek1 through threonine phosphorylation. Plant physiol. 2000, 122, 1301–1310. [Google Scholar] [CrossRef]
- Petersen, M.; Brodersen, P.; Naested, H.; Andreasson, E.; Lindhart, U.; Johansen, B.; Nielsen, H.B.; Lacy, M.; Austin, M.J.; Parker, J.E.; et al. Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 2000, 103, 1111–1120. [Google Scholar] [CrossRef]
- Suarez-Rodriguez, M.C.; Adams-Phillips, L.; Liu, Y.; Wang, H.; Su, S.H.; Jester, P.J.; Zhang, S.; Bent, A.F.; Krysan, P.J. Mekk1 is required for flg22-induced mpk4 activation in arabidopsis plants. Plant Physiol. 2007, 143, 661–669. [Google Scholar] [CrossRef]
- Li, B.; Jiang, S.; Yu, X.; Cheng, C.; Chen, S.; Cheng, Y.; Yuan, J.S.; Jiang, D.; He, P.; Shan, L. Phosphorylation of trihelix transcriptional repressor asr3 by map kinase4 negatively regulates arabidopsis immunity. Plant Cell 2015, 27, 839–856. [Google Scholar] [CrossRef]
- Sun, T.; Nitta, Y.; Zhang, Q.; Wu, D.; Tian, H.; Lee, J.S.; Zhang, Y. Antagonistic interactions between two map kinase cascades in plant development and immune signaling. EMBO Rep. 2018, 19, e45324. [Google Scholar] [CrossRef]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. Map kinase signalling cascade in arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, Y.; Huang, H.; Gao, M.; Wu, D.; Kong, Q.; Zhang, Y. The nlr protein summ2 senses the disruption of an immune signaling map kinase cascade via crck3. EMBO Rep. 2017, 18, 292–302. [Google Scholar] [CrossRef]
- Lumbreras, V.; Vilela, B.; Irar, S.; Sole, M.; Capellades, M.; Valls, M.; Coca, M.; Pages, M. Mapk phosphatase mkp2 mediates disease responses in arabidopsis and functionally interacts with mpk3 and mpk6. Plant J. Cell Mol. Biol. 2010, 63, 1017–1030. [Google Scholar] [CrossRef]
- Jiang, L.; Anderson, J.C.; Gonzalez Besteiro, M.A.; Peck, S.C. Phosphorylation of arabidopsis map kinase phosphatase 1 (mkp1) is required for pamp responses and resistance against bacteria. Plant Physiol. 2017, 175, 1839–1852. [Google Scholar] [CrossRef]
- Bartels, S.; Gonzalez Besteiro, M.A.; Lang, D.; Ulm, R. Emerging functions for plant map kinase phosphatases. Trends Plant Sci. 2010, 15, 322–329. [Google Scholar] [CrossRef]
- Fuchs, S.; Grill, E.; Meskiene, I.; Schweighofer, A. Type 2c protein phosphatases in plants. FEBS J. 2013, 280, 681–693. [Google Scholar] [CrossRef]
- Cui, F.; Sun, W.; Kong, X. Rlcks bridge plant immune receptors and mapk cascades. Trends Plant Sci. 2018, 23, 1039–1041. [Google Scholar] [CrossRef]
- Shinya, T.; Yamaguchi, K.; Desaki, Y.; Yamada, K.; Narisawa, T.; Kobayashi, Y.; Maeda, K.; Suzuki, M.; Tanimoto, T.; Takeda, J.; et al. Selective regulation of the chitin-induced defense response by the arabidopsis receptor-like cytoplasmic kinase pbl27. Plant J. Cell Mol. Biol. 2014, 79, 56–66. [Google Scholar] [CrossRef]
- Liu, T.; Liu, Z.; Song, C.; Hu, Y.; Han, Z.; She, J.; Fan, F.; Wang, J.; Jin, C.; Chang, J.; et al. Chitin-induced dimerization activates a plant immune receptor. Science 2012, 336, 1160–1164. [Google Scholar] [CrossRef]
- Bi, G.; Zhou, Z.; Wang, W.; Li, L.; Rao, S.; Wu, Y.; Zhang, X.; Menke, F.L.H.; Chen, S.; Zhou, J.M. Receptor-like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in arabidopsis. Plant Cell 2018, 30, 1543–1561. [Google Scholar] [CrossRef]
- Yamada, K.; Yamaguchi, K.; Yoshimura, S.; Terauchi, A.; Kawasaki, T. Conservation of chitin-induced mapk signaling pathways in rice and arabidopsis. Plant Cell Physiol. 2017, 58, 993–1002. [Google Scholar] [CrossRef]
- Wang, C.; Wang, G.; Zhang, C.; Zhu, P.; Dai, H.; Yu, N.; He, Z.; Xu, L.; Wang, E. Oscerk1-mediated chitin perception and immune signaling requires receptor-like cytoplasmic kinase 185 to activate an mapk cascade in rice. Mol. Plant 2017, 10, 619–633. [Google Scholar] [CrossRef]
- Tsuda, K.; Somssich, I.E. Transcriptional networks in plant immunity. New Phytol. 2015, 206, 932–947. [Google Scholar] [CrossRef]
- Allen, M.D.; Yamasaki, K.; Ohme-Takagi, M.; Tateno, M.; Suzuki, M. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the gcc-box binding domain in complex with DNA. EMBO J. 1998, 17, 5484–5496. [Google Scholar] [CrossRef]
- Li, X.; Duan, X.; Jiang, H.; Sun, Y.; Tang, Y.; Yuan, Z.; Guo, J.; Liang, W.; Chen, L.; Yin, J.; et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and arabidopsis. Plant Physiol. 2006, 141, 1167–1184. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. Myb transcription factors in arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of nac family genes in oryza sativa and arabidopsis thaliana. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. Wrky transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Droge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F.; bZIP Research Group. Bzip transcription factors in arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Vailleau, F.; Daniel, X.; Tronchet, M.; Montillet, J.L.; Triantaphylides, C.; Roby, D. A r2r3-myb gene, atmyb30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc. Natl. Acad. Sci. USA 2002, 99, 10179–10184. [Google Scholar] [CrossRef]
- Marino, D.; Froidure, S.; Canonne, J.; Khaled, S.B.; Khafif, M.; Pouzet, C.; Jauneau, A.; Roby, D.; Rivas, S. Addendum: Arabidopsis ubiquitin ligase miel1 mediates degradation of the transcription factor myb30 weakening plant defence. Nat. Commun. 2019, 10, 1475. [Google Scholar] [CrossRef]
- Marino, D.; Froidure, S.; Canonne, J.; Ben Khaled, S.; Khafif, M.; Pouzet, C.; Jauneau, A.; Roby, D.; Rivas, S. Arabidopsis ubiquitin ligase miel1 mediates degradation of the transcription factor myb30 weakening plant defence. Nat. Commun. 2013, 4, 1476. [Google Scholar] [CrossRef]
- Cheng, M.C.; Hsieh, E.J.; Chen, J.H.; Chen, H.Y.; Lin, T.P. Correction. Arabidopsis rglg2, functioning as a ring e3 ligase, interacts with aterf53 and negatively regulates the plant drought stress response. Plant Physiol. 2016, 170, 1162–1163. [Google Scholar]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the wrky53 transcription factor and its role during leaf senescence in arabidopsis. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef]
- Miao, Y.; Zentgraf, U. A hect e3 ubiquitin ligase negatively regulates arabidopsis leaf senescence through degradation of the transcription factor wrky53. Plant J. Cell Mol. Biol. 2010, 63, 179–188. [Google Scholar] [CrossRef]
- Matsushita, A.; Inoue, H.; Goto, S.; Nakayama, A.; Sugano, S.; Hayashi, N.; Takatsuji, H. Nuclear ubiquitin proteasome degradation affects wrky45 function in the rice defense program. Plant J. Cell Mol. Biol. 2013, 73, 302–313. [Google Scholar] [CrossRef]
- Choi, C.; Hwang, S.H.; Fang, I.R.; Kwon, S.I.; Park, S.R.; Ahn, I.; Kim, J.B.; Hwang, D.J. Molecular characterization of oryza sativa wrky6, which binds to w-box-like element 1 of the oryza sativa pathogenesis-related (pr) 10a promoter and confers reduced susceptibility to pathogens. New Phytol. 2015, 208, 846–859. [Google Scholar] [CrossRef]
- Lee, H.; Cha, J.; Choi, C.; Choi, N.; Ji, H.S.; Park, S.R.; Lee, S.; Hwang, D.J. Rice wrky11 plays a role in pathogen defense and drought tolerance. Rice 2018, 11, 5. [Google Scholar] [CrossRef]
- Mao, G.; Meng, X.; Liu, Y.; Zheng, Z.; Chen, Z.; Zhang, S. Phosphorylation of a wrky transcription factor by two pathogen-responsive mapks drives phytoalexin biosynthesis in arabidopsis. Plant Cell 2011, 23, 1639–1653. [Google Scholar] [CrossRef]
- Andreasson, E.; Jenkins, T.; Brodersen, P.; Thorgrimsen, S.; Petersen, N.H.; Zhu, S.; Qiu, J.L.; Micheelsen, P.; Rocher, A.; Petersen, M.; et al. The map kinase substrate mks1 is a regulator of plant defense responses. EMBO J. 2005, 24, 2579–2589. [Google Scholar] [CrossRef]
- Qiu, J.L.; Fiil, B.K.; Petersen, K.; Nielsen, H.B.; Botanga, C.J.; Thorgrimsen, S.; Palma, K.; Suarez-Rodriguez, M.C.; Sandbech-Clausen, S.; Lichota, J.; et al. Arabidopsis map kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 2008, 27, 2214–2221. [Google Scholar] [CrossRef]
- Menke, F.L.; Kang, H.G.; Chen, Z.; Park, J.M.; Kumar, D.; Klessig, D.F. Tobacco transcription factor wrky1 is phosphorylated by the map kinase sipk and mediates hr-like cell death in tobacco. Mol. Plant Microbe Interact. 2005, 18, 1027–1034. [Google Scholar] [CrossRef]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of osspl14 by osmir156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Withers, J.; Dong, X. Posttranslational modifications of npr1: A single protein playing multiple roles in plant immunity and physiology. PLoS Pathog. 2016, 12, e1005707. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, S.; Mou, Z. Nuclear localization of npr1 is required for regulation of salicylate tolerance, isochorismate synthase 1 expression and salicylate accumulation in arabidopsis. J. Plant Physiol. 2010, 167, 144–148. [Google Scholar] [CrossRef]
- Kinkema, M.; Fan, W.; Dong, X. Nuclear localization of npr1 is required for activation of pr gene expression. Plant Cell 2000, 12, 2339–2350. [Google Scholar] [CrossRef]
- Mou, Z.; Fan, W.; Dong, X. Inducers of plant systemic acquired resistance regulate npr1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N.; et al. Npr3 and npr4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef]
- Spoel, S.H.; Mou, Z.; Tada, Y.; Spivey, N.W.; Genschik, P.; Dong, X. Proteasome-mediated turnover of the transcription coactivator npr1 plays dual roles in regulating plant immunity. Cell 2009, 137, 860–872. [Google Scholar] [CrossRef]
- Saleh, A.; Withers, J.; Mohan, R.; Marques, J.; Gu, Y.; Yan, S.; Zavaliev, R.; Nomoto, M.; Tada, Y.; Dong, X. Posttranslational modifications of the master transcriptional regulator npr1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 2015, 18, 169–182. [Google Scholar] [CrossRef]
- Wang, D.; Amornsiripanitch, N.; Dong, X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006, 2, e123. [Google Scholar] [CrossRef]
- Despres, C.; DeLong, C.; Glaze, S.; Liu, E.; Fobert, P.R. The arabidopsis npr1/nim1 protein enhances the DNA binding activity of a subgroup of the tga family of bzip transcription factors. Plant Cell 2000, 12, 279–290. [Google Scholar] [CrossRef]
- Ning, Y.; Liu, W.; Wang, G.L. Balancing immunity and yield in crop plants. Trends Plant Sci. 2017, 22, 1069–1079. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, L.; Shi, H.; Chern, M.; Yu, H.; Yi, H.; He, M.; Yin, J.; Zhu, X.; Li, Y.; et al. A single transcription factor promotes both yield and immunity in rice. Science 2018, 361, 1026–1028. [Google Scholar] [CrossRef] [Green Version]


| TF Families | Descriptions |
|---|---|
| AP2/ERF | The proteins in this family contain an AP2/ERF DNA-binding domain that consists of three β-sheet strands followed by an α-helix motif [106]. |
| bHLH | This family is characterized by a basic-helix-loop-helix domain containing an N-terminal basic DNA-binding region and a C-terminal protein-interaction domain [107]. |
| MYB | This family is characterized by the presence of four repeat sequences, each containing three α-helices [108] |
| NAC | NAC TFs each contain a conserved DNA-binding domain on their N-termini and an activation domain on their C-termini [109]. |
| WRKY | The WRKY TFs contain WRKY domains having the typical WRKYGQK sequence followed by a zinc finger motif [110]. |
| bZIP | The bZIP proteins contain a basic region for DNA binding and a leucine zipper region for protein dimerization [111] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, J.; Yi, H.; Chen, X.; Wang, J. Post-Translational Modifications of Proteins Have Versatile Roles in Regulating Plant Immune Responses. Int. J. Mol. Sci. 2019, 20, 2807. https://doi.org/10.3390/ijms20112807
Yin J, Yi H, Chen X, Wang J. Post-Translational Modifications of Proteins Have Versatile Roles in Regulating Plant Immune Responses. International Journal of Molecular Sciences. 2019; 20(11):2807. https://doi.org/10.3390/ijms20112807
Chicago/Turabian StyleYin, Junjie, Hong Yi, Xuewei Chen, and Jing Wang. 2019. "Post-Translational Modifications of Proteins Have Versatile Roles in Regulating Plant Immune Responses" International Journal of Molecular Sciences 20, no. 11: 2807. https://doi.org/10.3390/ijms20112807




